1. Background
Staphylococcus aureus (S. aureus) is a highly virulent human pathogen and common cause of nosocomial and community-acquired infections (1). It is also a pathogen for many animal species, including intensively farmed food-producing animals. Worldwide, the dramatic increase in the prevalence of antimicrobial-resistant S. aureus (2-4) is receiving widespread attention, due to multi-resistant strains, diminishing the usefulness of antibiotics in human medicine and, thereby, limiting therapeutic options (5). Staphylococci are some of the most common causes of infections in birds (6). Most infections are caused by coagulase positive staphylococci, especially S. aureus, even though coagulase negative staphylococci also seem to be associated with infections (6-8).
Antimicrobial agents are widely used in the treatment and control of staphylococcal infections (9). S. aureus strains are able to acquire resistance to a variety of antimicrobial agents, such as oxacillin. Methicillin was first used for the treatment of S. aureus infections in 1959, and the first methicillin resistant S. aureus (MRSA) strain was reported in 1961 (10). Staphylococcal cassette chromosome mec (SCCmec), which is responsible for resistance to methicillin, is consisted of regulatory and structural genes. At the moment, 11 SCCmec types have been reported and are used for the typing of MRSA strains (11). It is suggested that extensive and, often, indiscriminate use of antimicrobial agents, in animal husbandry, might constitute a risk factor in creating an animal reservoir of antimicrobial-resistant bacteria (2, 12). From this reservoir, resistant strains or resistance genes might spread to humans, via the food chain. Data reports have indicated that food-producing animal species, raw poultry and other meat products harbor antimicrobial-resistant, Gram-positive bacteria (2, 12-14). However, only a few studies have determined the occurrence of antimicrobial resistance and presence of resistance genes, among staphylococci isolated from poultry (6, 14, 15).
2. Objectives
In this experimental study, we characterized the distribution and antibiotic resistance patterns of MRSA strains, isolated from lying hen farms in Iran. The pulsed field gel electrophoresis (PFGE) patterns and the SCCmec types were also determined.
3. Materials and Methods
3.1. Sample Collection and Identification
3.1.1. Swab Samples
Sampling was carried out during 2013, over a period of 90 days (collected on days 0, 45, 90) from three conventional laying hen farms, located in Karaj, Iran. All lying hens, which exhibited no signs of clinical disease, were selected, with ages ranging from 20 - 24 weeks. Nine samplings were done from three poultry farms (three each) using sterile cotton swabs from the cloaca of each chicken. Swabs were aseptically transferred to brain heart infusion broth, supplemented with 10 µg/mL of colistin and nalidixic acid and incubated for 24 hours, at 37°C. Ten microliter of each sample were streaked on chromogenic MRSA selective agar [Oxoid Brilliance MRSA Agar (Thermo Fisher Scientific, Waltham, MA, USA)] and incubated for 48 hours, at 37°C.
3.1.2. Litter Collection
Litter samples were collected over a period of 90 days (collected ondays 0, 45, 90) in 2013, from three conventional laying hen farms. At each farm, the waste material was collected from top 25 - 50 cm of poultry litter, from the poultry house floor, and transferred to 500ml sterile bottles. Samples were analyzed within 24 hours of collection. All three farmers reported that no recognized disease outbreaks had occurred during the flock cycle, so that no therapeutic drug use was applied. However, no specific information on antibiotic feed additives was available from the producers, as this is considered confidential business information (16).
Each chicken litter sample was mixed in the 500 mL sterile bottle, by vigorously agitating the bottle by hand for 1 minute. Five grams of litter were then placed in 45 mL of phosphate-buffered saline and were diluted five folds, before filtration on a 0.45-µm membrane (Millipore Corporation, Bedford, MA, USA) (16). Filter membrane, transferred to chromogenic MRSA selective agar [Oxoid Brilliance MRSA Agar (Thermo Fisher Scientific, Waltham, MA, USA)] and incubated for 48 hours, at 37°C.
All colonies cultured on blood agar plates (Merck, Darmstadt, Germany) and pure culture were initially identified to the species level, using nucA specific primers. A High Pure PCR Template Preparation kit (Roche, Mannheim, Germany) was employed for DNA extraction of strains, according to the instructions of the manufacturer. Different primers used in this study are listed in Table 1. Also, polymerase chain reaction (PCR) and Multiplex-PCR were mixed in a volume of 25 μL consisting of 1 µL of template DNA, 10X PCR buffer, dNTP mix (100 μM), MgCl2 (0.8 μM), each primer (0.4 μM) and taq DNA polymerase (1 U).
| Gene | Sequence | Molecular Weight, bp | Reference |
|---|---|---|---|
| nucA | 400 | (17) | |
| Forward | 5’-AGTTCAGCAAATGCATCACA | ||
| Reverse | 5’-TAGCCAAGCCTTGACGAACT | ||
| mecA | 310 | (18) | |
| Forward | 5’-GTAGAAATGACTGAACGTCCGATAA | ||
| Reverse | 5’-CCAATTCCACATTGTTTCGGTCTAA | ||
| SCCmec Type I | 613 | (19) | |
| Forward | 5’-GCTTTAAAGAGTGTCGTTACAGG | ||
| Reverse | 5’-GTTCTCTCATAGTATGACGTCC | ||
| SCCmec Type II | 398 | (19) | |
| Forward | 5’-CGTTGAAGATGATGAAGCG | ||
| Reverse | 5’-CGAAATCAATGGTTAATGGACC | ||
| SCCmec Type III | 280 | (19) | |
| Forward | 5’-CCATATTGTGTACGATGCG | ||
| Reverse | 5’-CCTTAGTTGTCGTAACAGATCG | ||
| SCCmec Type IVa | 776 | (19) | |
| Forward | 5’-GCCTTATTCGAAGAAACCG | ||
| Reverse | 5’-CTACTCTTCTGAAAAGCGTCG | ||
| SCCmec Type IVb | 493 | (19) | |
| Forward | 5’-TCTGGAATTACTTCAGCTGC | ||
| Reverse | 5’-AAACAATATTGCTCTCCCTC | ||
| SCCmec Type IVc | 200 | (19) | |
| Forward | 5’-ACAATATTTGTATTATCGGAGAGC | ||
| Reverse | 5’-TTGGTATGAGGTATTGCTGG | ||
| SCCmec Type IVd | 881 | (19) | |
| Forward | 5’-CTCAAAATACGGACCCCAATACA | ||
| Reverse | 5’-TGCTCCAGTAATTGCTAAAG | ||
| SCCmec Type V | 325 | (19) | |
| Forward | 5’-GAACATTGTTACTTAAATGAGCG | ||
| Reverse | 5’-TGAAAGTTGTACCCTTGACACC | ||
| Type 1 - 3 ccr | ccrAB (forward): 5’-ATTGCCTTGATAATAGCCITCT | (19) | |
| Type 1 ccr | ccrA1B1 (reverse): 5’-AACCTATATCATCAATCAGTACGT | 700 | (19) |
| Type 2 ccr | ccrA2B2 (reverse): 5’-TAAAGGCATCAATGCACAAACACT | 1000 | (19) |
| Type 3 ccr | ccrA3B3(reverse): 5’-AGCTCAAAAGCAAGCAATAGAAT | 1600 | (19) |
| pvl | 433 | (18) | |
| Forward | 5’-ATCATTAGGTAAAATGTCTGGACATGATCCA | ||
| Reverse | 5’-GCATCAAGTGTATTGGATAGCAAAAGC |
The PCR primers specific for nucA (17) (cycle conditions: initial activation at 94°C for 5 minute; 30 cycles at 94°C for 45 seconds, 62°C for 45 seconds, 72°C for 105 seconds; final extension at 72°C for 10 minutes) and methicillin resistance genes (mecA) (18) (cycle conditions: initial activation at 94°C for 10 minutes; 25 cycles at 94°C for 45 seconds, 55°C for 45 seconds, 72°C for 75 seconds; final extension at 72°C for 10 minutes) were synthesized by TIB Molbiol Syntheselabor GmbH (Berlin, Germany).
3.2. Antibiotic Susceptibility Tests
All strains were tested for susceptibility to cefoxitin (30 µg), penicillin (10 IU), gentamicin (10 µg), amikacin (30 µg), kanamycin (30 µg), tobramycin (10 µg), tetracycline (30 µg), minocycline (30 µg), clindamycin (2 µg), erythromycin (15 µg), quinupristin-dalfopristin (15 µg), linezolid (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), nitrofurantoin (300 µg), rifampin (5 µg), and trimethoprim-sulphamethoxazole (1.25 - 23.75 µg), according to the guidelines of Clinical and Laboratory Standards Institute (20). The antibiotic discs were purchased from Mast Diagnostics (Merseyside, United Kingdom). The minimal inhibitory concentrations (MICs) for oxacillin and vancomycin of MRSA strains were determined using broth microdilution assay (21).
3.3. Typing of Methicillin-Resistant Staphylococcus aureus Isolates
3.3.1. Pulsed Field Gel Electrophoresis
All MRSA isolates were typed by PFGE assay, according to the protocol described previously by Chung et al. (22). Salmonella choleraesuis serotype Branderup H9812 was included, as a molecular size marker. The unweighted pair group method with arithmetic mean method, using Gelcompare II software version 4.0 (Applied Maths, Sint-Matens-Latem, Belgium), was employed for analysis of banding patterns of MRSA strains (23).
3.3.2. SCCmec and ccr Typing
The SCCmec typing of MRSA strains was done using a multiplex PCR typing assay, containing eight pairs of primers, including the unique and specific primers for SCCmec types and subtypes I, II, III, IVa, IVb, IVc, IVd, and V (cycle conditions: initial activation at 94°C for 5 minutes; 10 cycles at 94°C for 45 seconds, 65°C for 45 seconds and 72°C for 1.5 minutes; 25 cycles at 94°C for 45 seconds, 55°C for 45 seconds and 72°C for 1.5 minutes; a final extension at 72°C for 10 minutes) (19). Another multiplex PCR assay was used for characterization of ccr gene complexes, by the cycle conditions described previously for SCCmec typing (19).
3.4. Detection of pvl Gene
The pvl gene, encoding Panton-Valentine leukocidin (PVL) was detected using specific primers (18) and PCR cycles described previously (cycle conditions: initial activation at 94°C for 10 minutes; 10 cycles at 94°C for 45 seconds, 55°C for 45 seconds and 72°C for 75 seconds; 25 cycles at 94°C for 45 seconds, 50°C for 45 seconds, and 72°C for 75 seconds; a final extension at 72°C for 10 minutes) (10).
4. Results
4.1. Identification of Isolates
A total of 55 suspected colonies isolates were included in this study. The PCR, using nuc primers, confirmed all isolates as S. aureus. All strains were resistant to methicillin and cefoxitin, harbored mecA gene and identified as MRSA.
4.2. Antibiotic Susceptibility Testing
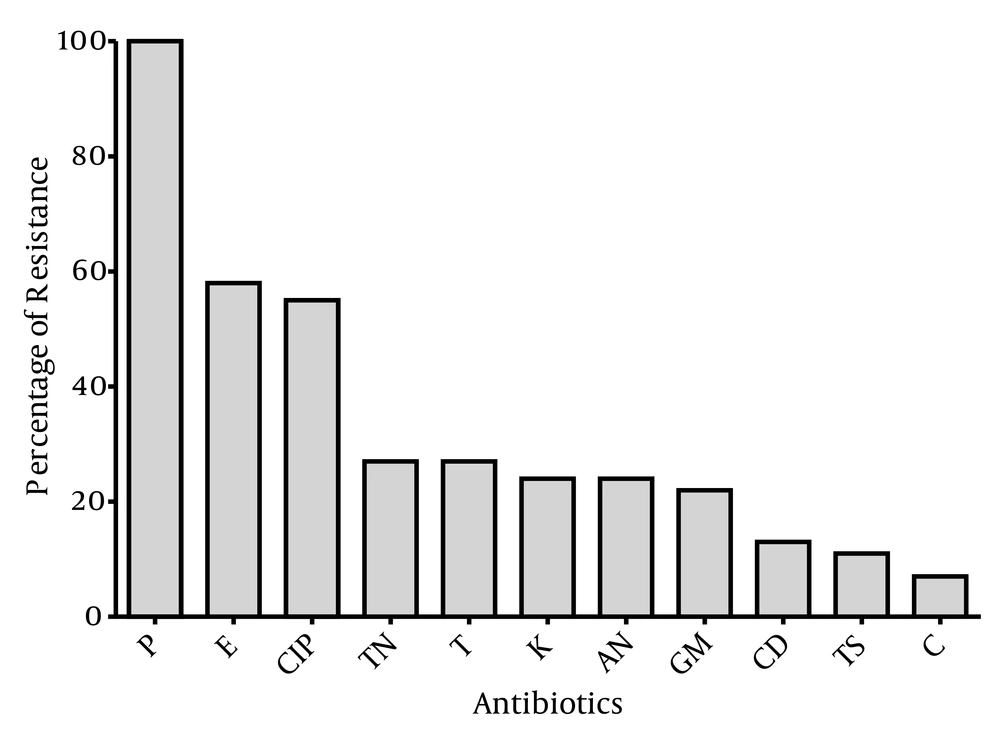
The susceptibility of the 55 MRSA isolates to antimicrobial agents is shown in Figure 1. The MRSA isolates showed wide range of resistance to different antibiotics and 100% of MRSA strains were resistant to penicillin. Moreover, all isolates were susceptible to vancomycin, linezolid, quinupristin-dalfopristin, minocycline, nitrofurantoin and fusidic acid. Resistance to erythromycin and ciprofloxacin was observed among 58% and 55% of strains, respectively.
A total of 16 antibiotic resistance patterns were seen among MRSA isolates, when bacteria were grouped according to ten antibiotics (Table 2), in which 18 strains (32.7%) were susceptible to all classes of antibiotics tested, except for penicillin. Moreover, most of the isolates (16.4%) showed resistance to three antibiotics (patterns 4 - 6) and seven isolates (12.7%) were resistant to eight different antibiotics (patterns 14 - 16).
| No. of strains resistant to: | No of Farm 1 Isolates | No of Farm 2 Isolates | No of Farm 3 Isolates | Frequency |
|---|---|---|---|---|
| One antibiotic | 8 (36.3) | 5 (29.4) | 5 (31.3) | 18 (32.7) |
| P | 8 (36.3) | 5 (29.4) | 5 (31.3) | 18 (32.7) |
| Two antibiotics | 2 (9.1) | 2 (11.8) | 1 (6.3) | 5 (9.1) |
| P, CIP | 1 (4.5) | 2 (11.8) | 0 | 3 (5.5) |
| P, E | 1 (4.5) | 0 | 1 (6.3) | 2 (3.6) |
| Three antibiotics | 3 (13.6) | 4 (23.5) | 2 (12.5) | 9 (16.4) |
| P, CIP, E | 2 (9.1) | 1 (5.9) | 1 (6.3) | 4 (7.3) |
| P, E, T | 1 (4.5) | 2 (11.8) | 0 | 3 (5.5) |
| P, CIP, T | 0 | 1 (5.9) | 1 (6.3) | 2 (3.6) |
| Four antibiotics | 1 (4.5) | 1 (5.9) | 1 (6.3) | 3 (5.5) |
| P, CIP, E, T | 1 (4.5) | 1 (5.9) | 1 (6.3) | 3 (5.5) |
| Five antibiotics | 1 (4.5) | 1 (5.9) | 2 (12.5) | 4 (7.3) |
| P, CIP, E, T, CD | 1 (4.5) | 0 | 1 (6.3) | 2 (3.6) |
| P, E, K, AN, TN | 0 | 1 (5.9) | 1 (6.3) | 2 (3.6) |
| Six antibiotics | 2 (9.1) | 1 (5.9) | 1 (6.3) | 4 (7.3) |
| P, CIP, E, T, CD, C | 1 (4.5) | 0 | 0 | 1 (1.8) |
| P, CIP, E, K, AN, TN | 1 (4.5) | 1 (5.9) | 1 (6.3) | 3 (5.5) |
| Seven antibiotics | 3 (13.6) | 1 (5.9) | 1 (6.3) | 5 (9.1) |
| P, CIP, E, K, AN, TN, GM | 2 (9.1) | 1 (5.9) | 0 | 3 (5.5) |
| P, CIP, E, T, CD, TS, GM | 1 () | 0 | 1 (6.3) | 2 (3.6) |
| Eight antibiotics | 2 (9.1) | 2 (11.8) | 3 (18.8) | 7 (12.7) |
| P, CIP, E, K, AN, TN, GM, C | 1 (4.5) | 0 | 2 (12.5) | 3 (5.5) |
| P, CIP, E, K, AN, TN, GM, TS | 0 | 2 (11.8) | 0 | 2 (3.6) |
| P, CIP, E, T, CD, TS, GM, TN | 1 (4.5) | 0 | 1 (6.3) | 2 (3.6) |
aAbbreviations: AN, Amikacin; C, Chloramphenicol; CIP, Ciprofloxacin; CD, Clindamycin; E, Erythromycin; GM, Gentamicin; K, Kanamycin; P, Penicillin; T, Tetracycline; TN, Tobramycin; TS, Trimethoprim-Sulphamethoxazole.
bValues are presented as No. (%).
4.3. Typing of Methicillin-Resistant Staphylococcus aureus Isolates
4.3.1. Pulsed-Field gel Electrophoresis
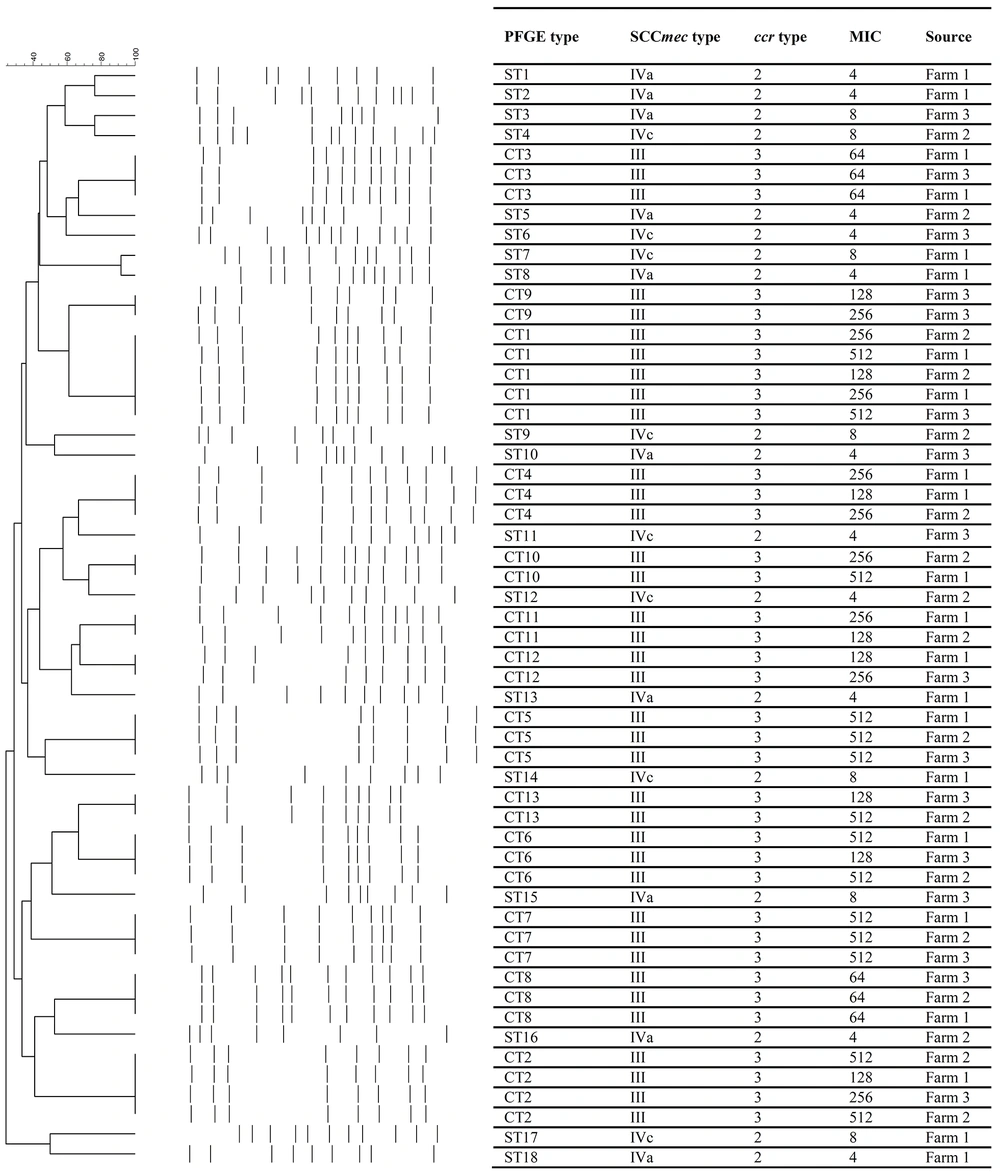
The results of PFGE typing for the MRSA isolates discriminated 55 strains to 31 pulsotypes, consisting of 18 single types (ST1 - ST18) and 13 common types (CT1 - CT13) (Figure 2). The presence of different STs was limited to the isolates harboring SCCmec type IV (a or c) and pvl gene. Such isolates showed the highest susceptibility to all antibiotics tested, except for penicillin. On the other hand, CT1 was the dominant pulsotype, consisting of five strains (9.1%). All MRSA strains in different CTs (n = 105) harbored SCCmec type III and carried ccr type 3. The CTs 1, 2 and 4 - 8 were common among all three laying farms and CT3 was only found in farm three.
4.3.2. SCCmec and ccr Typing
Two different ccr types and three SCCmec types were detected among MRSA isolates (Figure 2). SCCmec type IVa and IVc were present in 10 (18%) and eight (15%) MRSA strains, respectively. On the other hand, 37 (67%) isolates were positive for SCCmec type III, and classified as hospital acquired MRSA (HA-MRSA) strains. Moreover, 33% and 67% of the isolates harbored ccr type 2 and 3, respectively.
4.4. Detection of pvl Gene
In this study, 18 strains (67%) were positive for pvl gene (Figure 2). The presence of pvl gene was limited to the strains harboring SCCmec type IV and also showed low level of resistance to oxacillin.
The results of MIC of oxacillin showed that all of the isolates were resistant to oxacillin (MIC ≥ 4 µg/mL) (Figure 2). Twenty percent and 41.4% of the MRSA isolates showed low (MIC ≥ 4 µg/mL) and high (MIC ≥ 256 µg/mL) resistance to oxacillin, respectively.
5. Discussion
In this study, we could isolate MRSA isolates from laying hens in each farm and in each sampling. This finding indicates that MRSA may persist on a farm and colonize future flocks. There are different reports, from around the world, with different frequencies. Persons and colleagues, in Belgium (15), reported 11% the prevalence of MRSA in broiler chickens; however, they could not isolate any MRSA isolate from laying hens. On the other hand, a low prevalence of MRSA in poultries has also been found by other investigators (24, 25), although they sampled chicken carcasses from slaughterhouses and did not find any livestock-associated strains.
The occurrence of erythromycin resistance among poultry MRSA was significantly lower than that observed among MRSA of human origin, in Iran (10, 26-29). Macrolides are not normally used for therapy of infections in poultry in Denmark, whereas the frequent occurrence of macrolide resistance might reflect the use of the spiramycin, as a growth promoter of poultry, in Iran. This finding is in contrast to a report of another study, in Belgium, that showed all MRSA strains, isolated in that study, were resistant to erythromycin (15). Also, Aarestrup and colleagues, in Denmark, showed that only 24% of the isolates were resistant to this antibiotic (6). Differences in frequency of resistance to erythromycin could result from the differences in patterns of antibiotic consumption in their country and use of antibiotic, as a growth promoter of poultry.
The frequency of penicillin resistance in this study was high and 100% of MRSA isolates showed resistance to this antibiotic. In other studies from Iran, the prevalence of penicillin resistance among MRSA isolates was 100% (10, 26-28, 30). Possibly, the general use of ampicillin for the treatment of infections in poultry may explain this finding.
These findings showed a very frequent occurrence of resistance to tetracycline among MRSA isolates. This rate of resistance is similar to other studies in the world (2, 6, 31) and lower than another study in Belgium (15). It might be due to high consumption of tetracycline for treatment of S. aureus infections, in Iran.
The MRSA was isolated from any laying hen samples. This finding may indicate that MRSA is present in high numbers, in laying hens, possibly because of high use of antimicrobial drugs in these animals. Use of certain antimicrobial drugs in human hospitals has been shown to be a risk factor for acquiring MRSA infection, especially when the chosen treatment is inappropriate or insufficient (15). Antimicrobial-drug use may also be a risk factor for MRSA colonization of animals. The antimicrobial drugs, used in Iran, are macrolides, colistin, amoxicillin, tetracycline and trimethoprim-sulfamethoxazole.
Also, for new generation antibiotics, such as linezolid or quinupristin-dalfopristin, no resistance has been reported by investigators in Iran (2, 6, 15, 31). In addition, even with the high usage of vancomycin, no vancomycin resistant S. aureus (VRSA) or vancomycin intermediate S. aureus (VISA) were seen in this study, suggesting the increased use of certain antibiotics is not sufficient to ensure the appearance of the resistant strains and, other factors, such as environmental and genetic ones, must be considered.
In this study, the frequency of SCCmec type IV was 33%. As it has been shown (32), PVL is the marker of SCCmec type IV and encoded by SGA prophage type and the relation between presence of SGA prophage and SCCmec type IV was shown previously (10). In different studies in Iran, 9% and 6% of clinical and sewage origin isolates carried SCCmec type IV, respectively (10, 30). Moreover, high prevalence of SCCmec type III, among lying farms, indicated the hospital origin of strains. Previous reports in Iran revealed that SCCmec type III was the dominant type among isolates with hospital and environmental origin (10, 30, 33, 34). Rahimi et al. showed the epidemiological link between MRSA strains with sewage and hospital origin, in Tehran, Iran (30). These findings, altogether, reported that SCCmec type III is the dominant SCCmec type among MRSA strains with different origin, in Iran. On the other hand, Hauschild and colleagues reported SCCmec type IV and V as dominant types among poultry, in Germany (35).
The PFGE typing of MRSA strains showed a genetic diversity among MRSA isolated from different farms. The presence of some of the isolates, in all three farms (CT1, CT2 and CT4 - CT8), indicating widespread dissemination of these bacterial clones, which further supports the spread of such clonal types in lying farms tested. The predominant pulsotypes CT1 - CT3 and their dissemination in this study are consistent with previous results, indicating a wide distribution of specific clonal groups of MRSA in Tehran (10, 36). In a previous study, we showed that community acquired MRSA (CA-MRSA) strains were completely different from HA-MRSA strains and belonged to different clones. Such strains were only resistant to beta-lactam antibiotics, harbored pvl gene and were positive for SCCmec type IV and type 2 ccr (10). In contrast to another report from Tehran (36), strains with highly similar pulsotypes had different antibiotic resistance patterns, probably due to acquisition of new characteristics during their long residency in the environment.
In conclusion, the results of this study illustrate the persistence of resistant bacteria in the environment and highlight the reservoir of resistance associated with the use of antibiotics, as a feed additive in poultry production. Further, the presence of antibiotic resistant enteric bacteria, in the poultry production environment, increases the potential for human exposure to drug resistant bacteria.
Here we confirmed the presence of MRSA in laying hens in Iran. All isolates belonged to different antibiotic resistance patterns, and, therefore, were similar to other strains isolated from clinical samples in Iran and abroad. Whether the presence of SCCmec types III and IV is typically associated with poultry, still needs to be confirmed. More detailed data are also needed to gain further insight in the true within- and between flock prevalence of MRSA, in poultry, and its evolution over time. Better understanding of emergence and dissemination of MRSA, in different sources, requires the careful study of different various aspects of species and characterization of interspecies transmission.