1. Background
Staphylococcus epidermidis are a member of coagulase negative gram-positive bacteria, which are known as a common human microbiota and cause infections in hospitalized patients (1). Staphylococcus epidermidis are opportunistic pathogens, which lack virulence factors and are known as the most important cause of indwelling device-related infections (2).
Staphylococcus epidermidis has the ability to form biofilm, which consists of multilayered bacterial communities stabilized by intercellular adhesive mechanisms that do not respond to antimicrobial treatments (3). Staphylococcus epidermidis binds to extracellular matrix proteins via microbial surface components recognizing adhesive matrix molecules (MSCRAMMS) (4). Currently, the best understood biofilm mechanism in S. epidermidis is synthesis of major component of exopolysaccharide matrix, polysaccharide intercellular adhesion (PIA) or Polymeric N-Acetyl-Glucosamine (PNAG) (2, 5). Adhesion factors, such as intercellular adhesin locus (ica) composed of genes icaA, icaD, icaB and icaC, which code for polysaccharide intercellular adhesin (PIA) and biofilm associated-homologous protein (bhp) play an important role in the ability of S. epidermidis to persist and form multilayered bacterial communities (6, 7). It has been shown that prevalence of ica locus among clinical S. epidermidis strains isolated from catheter-related infections and sepsis is higher than other sources (8, 9). In strains that lack the ica locus, biofilm formation is due to the presence of aap gene, which enables bacteria to bind to various matrix proteins (5). Insertion of transposon IS256 into the ica locus results in change of biofilm formation and resistance to aminoglycosides, which converts biofilm positive to biofilm negative bacteria (9, 10).
Biofilm communities show high-level resistance to antimicrobial agents, which is due to reduced cell division, DNA replication and synthesis of different proteins (5). Moreover, biofilm results in recurrent infections, which are difficult to treat and lead to higher treatment costs (3).
2. Objectives
In this experimental study, we aimed to describe the antibiotic resistance patterns of biofilm-producing S. epidermidis strains isolated from clinical samples in Tehran, Iran. Moreover, the role of different genes in biofilm formation was also described.
3. Patients and Methods
3.1. Sample Collection and Identification
During February to December 2014, a total of 250 S. epidermidis strains were isolated from a private hospital in Tehran. Fifty percent, 32%, 16% and 2% of isolates were collected from blood, catheter, wound and eye culture, respectively. All isolates were cultured on blood agar medium (Merck KGaA, Darmstadt, Germany) and pure colonies were analyzed using biochemical tests such as oxidase, catalase, DNase, coagulase and mannitol fermentation tests (11). All identified isolates were subjected to PCR using species-specific gseA gene primers (12). DNA of suspected isolates was extracted by the boiling method, according to the protocol introduced previously by Rahimi et al. (13).
3.2. Biofilm Formation
3.2.1. Qualitative Method
Staphylococcus epidermidis isolates were tested for slime production on Congo-red (CR) agar palates (14). All isolates were cultured on CR agar plates and incubated for 24 hours at 37°C. Black and red colonies were classified as positive and negative for slime production, respectively. Staphylococcus epidermidis ATCC 35984 and S. epidermidis ATCC 12228, as positive and negative controls, were used in all qualitative tests.
3.2.2. Quantitative Method
To measure the adherence ability of bacteria, the quantitative crystal violet microtiter plate assay was employed according to the protocol of Wang et al. (15). Biofilm formation was classified as negative (OD570 ≤ ODc), weak (OD570 < 0.2), moderate (0.2 ≤ OD570 < 0.5) and strong (OD570 ≥ 0.5).
3.2.3. Antibiotic Susceptibility Tests
All S. epidermidis strains were tested for susceptibility to 17 antibiotics using the disk diffusion method by the guidelines of clinical laboratory and standard institute (CLSI) (16). Antibiotics used in this study were cefoxitin (30 µg), clindamycin (2 µg), erythromycin (15 µg), amikacin (30 µg), kanamycin (30 µg), gentamicin (10 µg), tobramycin (10 µg), tetracycline (30 µg) minocycline (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), trimethoprim-sulfamethoxazole (1.25 - 23.75 µg), linezolid (30 µg), rifampin (5 µg), nitrofurantoin (300 µg), penicillin (10 IU) and quinupristi-dalfopristin (15 µg) (Mast Diagnostics, Bootle, Mersey Side,. UK). The susceptibility of the strains to fusidic acid was determined using the disc diffusion method, according to Rahimi et al. (17). The minimum inhibitory concentration (MIC) of vancomycin and oxacillin was tested using the E test, according to the guidelines of the manufacturer (AB bioMerieux, Solna, Sweden).
3.2.4. DNA Extraction and Polymerase Chain Reaction
DNA of all biofilm-positive S. epidermidis strains was extracted using high pure PCR template preparation kit (Roche, Germany), according to the manufacturer’s recommendations. polymerase chain reactions were employed for detection of icaA, icaB, icaC, icaD, app, IS256, atlE and bhp genes by specific primers, as described previously (10, 18-21).
3.2.5. Statistical Analysis
To evaluate data for univariate comparisons of categorical results by Fisher’s exact test, GraphPad Prism 5.0 (GraphPad Software) was employed for statistical analysis.
4. Results
4.1. Identification of Bacteria
Using the PCR method by specific primers, all 250 isolates were confirmed as S. epidermidis strains. In this study, 32%, 30%, 28%, 4%, 4%, and 2% of strains were isolated from patients in the range of ≥ 51, 36 - 50, 20 - 35, 11 - 19, 2 - 10 and 1 ≤ years, respectively.
4.2. Biofilm Formation
4.2.1. Qualitative Method
Among 250 S. epidermidis strains isolated from different patients, 205 (82%) strains were able to produce black colonies, and were known as slime-producing strains. On the other hand, 45 (18%) strains produced dark-red to red colonies and were classified as biofilm-negative strains.
4.2.2. Quantitative Method
Among all 205 slime producing S. epidermidis strains tested for biofilm formation by quantitative method, 170 (68%), 21 (8%) and 14 (6%) strains exhibited high (OD570 ≥ 0.5), moderate (0.2 ≤ OD570 < 0.5) and weak (OD570 < 0.2) level of biofilm formation. Moreover, 45 non-slime producing strains (18%) were not able to form biofilm (OD570 ≤ ODc).
4.3. Antibiotic Susceptibility Tests
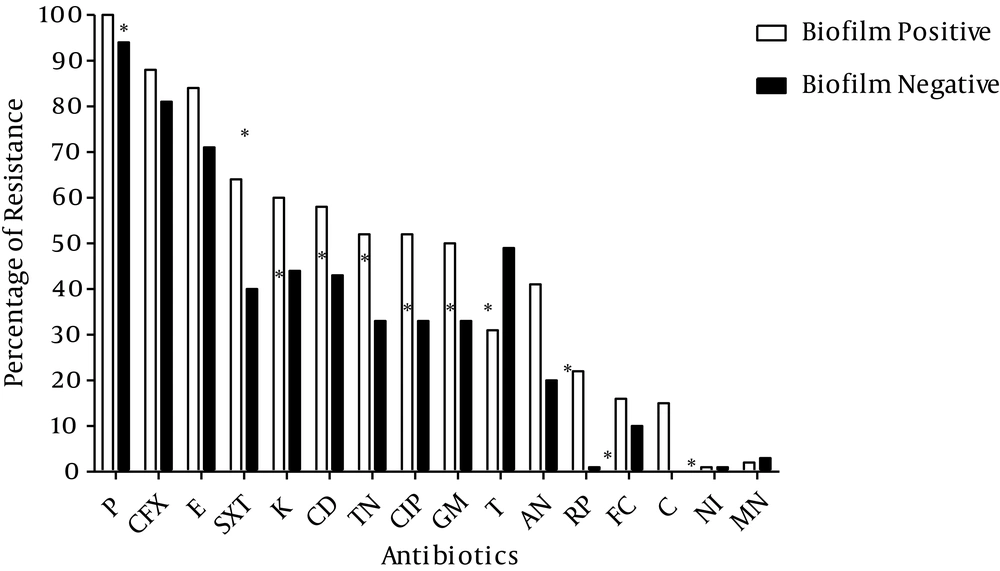
As shown in Figure 1, 86% of S. epidermidis strains showed resistance to cefoxitin and were classified as methicillin resistant S. epidermidis (MRSE). Moreover, 98% and 80% of strains showed resistance to penicillin and erythromycin, respectively. Furthermore, 56%, 54% and 53% of strains were resistant to trimethoprim-sulfamethoxazole, kanamycin and clindamycin, respectively. None of the isolates showed resistance to linezolid, quinupristin-dalfopristin and vancomycin and low level resistance to nitrofurantoin and minocycline was observed. On the other hand, the rate of resistance to different antibiotics (except for fusidic acid, nitrofurantoin and minocycline) was significantly higher among biofilm-producing strains compared to non-biofilm-producing isolates.
Amongst these isolates, 37 different antibiotic resistance patterns were identified (data not shown), and bacteria showed resistance to 1 - 13 antibiotics. Ten (4%) biofilm-producing and non-biofilm-producing S. epidermidis strains showed susceptibility to all tested antibiotics except for penicillin. Twenty-five (14.7%) and 26 (15.3%) biofilm-producing strains showed resistance to 13 and 5 antibiotics, respectively, whilst 17.5% and 13.8% of the non-biofilm producing strains showed resistance to five and seven antibiotics, respectively. Most of the biofilm-producing strains (49.4%) showed resistance to 9-13 antibiotics.
The results of MIC of 250 S. epidermidis strains to oxacillin revealed that 65.9% (112 isolates) of the biofilm-producing strains and 22.6% (18 isolates) of non-biofilm-producing strains showed high level of resistance (MIC ≥ 128 µg/mL) to oxacillin (Table 1). Five biofilm-producing strains showed low level of resistance to oxacillin (MIC ≥ 4 and 8 µg/mL), while five non-biofilm-producing strains were also resistant to low level (MIC ≥ 4 and 12 µg/mL) of oxacillin. These 10 strains showed susceptibility to all antibiotics except for penicillin.
| MIC, μg/mL | 256 | 192 | 128 | 96 | 64 | 48 | 32 | 24 | 16 | 12 | 8 | 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biofilm Producing | 24 (14.1) | 35 (20.6) | 53 (31.2) | 18 (10.6) | 11 (6.5) | 9 (5.3) | 4 (2.3) | 7 (4.1) | 4 (2.3) | - | 3 (1.8) | 2 (1.2) |
| Non-Biofilm Producing | 7 (8.8) | 3 (3.8) | 8 (10) | 21 (26.2) | 8 (10) | 17 (21.2) | 4 (5) | 3 (3.8) | 4 (5) | 4 (5) | - | 1 (1.2) |
| Total | 31 (12.4) | 38 (15.2) | 61 (24.4) | 39 (15.6) | 19 (7.6) | 26 (10.4) | 8 (3.2) | 10 (4) | 8 (3.2) | 4 (1.6) | 3 (1.2) | 3 (1.2) |
aValues are expressed as No. (%).
4.4. Detection of Genes Involved in Biofilm Formation
Amongst all S. epidermidis isolates, 195 (78%) and 180 (72%) strains were positive for aap and IS256 genes, respectively (Table 2). On the other hand, all 170 biofilm-producing strains harbored atlE, aap, icaA and icaD genes and 93%, 83% and 38% of strains were positive for icaB, icaC and bhp genes, respectively. Furthermore, in non-biofilm producing strains, 19%, 50%, 46%, and 40% of the isolates carried icaA, icaD, IS256 and atlE genes, respectively. The presence of all genes involved in biofilm formation among biofilm positive strains was significantly higher than non-biofilm-producing strains.
| Biofilm Genes | Biofilm Producing | Non-Biofilm Producing | Total | P Value |
|---|---|---|---|---|
| icaA | 170 (100) | 15 (19) | 185 (74) | < 0.0001 |
| icaB | 158 (93) | 12 (15) | 170 (68) | < 0.0001 |
| icaC | 141 (83) | 9 (11) | 150 (60) | < 0.0001 |
| icaD | 170 (100) | 40 (50) | 210 (84) | < 0.0001 |
| aap | 170 (100) | 25 (31) | 195 (78) | < 0.0001 |
| IS256 | 143 (84) | 37 (46) | 180 (72) | < 0.0001 |
| bhp | 65 (38) | 19 (24) | 84 (34) | 0.0309 |
| atlE | 170 (100) | 32 (40) | 202 (81) | < 0.0001 |
aValues are expressed as No. (%).
5. Discussion
We showed the high prevalence of biofilm-forming S. epidermidis strains isolated from hospitalized patients in a private hospital in Tehran, Iran, which most of them isolated from blood and catheter samples. It has been shown previously that catheters could be a risk factor for staphylococcal infections (1).
The results of crystal violet semi-quantitative and qualitative Congo Red agar assay were not consistent with each other. In this study, 82% of S. epidermidis strains were able to produce black colonies on Congo Red agar medium, this rate was higher than other reports (18, 22, 23). On the other hand, in a microtiter plate assay, 68% of strains were able to form biofilms; this rate of biofilm formation was lower than other studies (2, 10, 24). Overall, these findings are consistent with other studies (9, 25-28), but in Germany, results of qualitative and quantitative tests were the same (10, 24). These differences indicated that crystal violet semi-quantitative PCR could be the method of choice for screening and assay of biofilm formation in S. epidermidis strains.
In present study, we found high level resistance to penicillin, cefoxitin and erythromycin among biofilm and non-biofilm producing S. epidermidis strains, amongst these no significant difference was observed among biofilm and non-biofilm producing strains for resistance to cefoxitin. High level of resistance to such antibiotics could be due to the frequent use of these antibiotics in hospitals of Tehran. Low rate of resistance to some antibiotics such as ciprofloxacin, clindamycin, tobramycin, tetracycline and amikacin was surprising yet this might be due to the non-frequent use of such antibiotics for treatment of infections caused by S. epidermidis strains in Iran. Resistance to antibiotics and also production of biofilm are considered to be virulence factors that help the survival of bacteria in the host.
In this study, 86% of S. epidermidis strains were resistant to cefoxitin and classified as Methicillin-Resistant Staphylococcus epidermidis (MRSE) strains; these values are higher than other reports (10, 25, 29). The prevalence of erythromycin-resistant strains was significantly lower in other studies (30-32). On the other hand, we found that 10 biofilm and non-biofilm-producing strains showed susceptibility to all tested antibiotics except for penicillin; these findings are consistent with our previous reports for S. aureus strains (17, 33-38). Moreover, resistance of biofilm-producing strains to 13 tested antibiotics in this study highlighted the important role of these strains to cause severe infections, which do not respond to broad spectrum of antibiotics.
All biofilm-producing S. epidermidis strains were positive for icaA, icaD, aap and atlE genes. This finding is in contrast to a previous study, where these genes were not detected in all biofilm-producing isolates (18) and is in agreement with other reports (23, 25-27, 39). Comparison of biofilm-forming and non-biofilm-producing strains may indicate the importance and necessity of ica genes for slime layer production and biofilm formation, which is consistent with other reports (4, 23, 39), but it is necessary to investigate the expression of icaA and icaD genes using mutant strains to conclude role of these genes on biofilm formation. Different studies revealed the importance of the aap gene for attachment of S. epidermidis strains and presence of icaA and icaD, together with aap and atlE genes among S. epidermidis strains indicating the high potential of such strains to attach and form biofilms. It has been shown that, these genes are markers for clinically relevant strains (1).
The presence of IS256 transposon in ica locus has been reported previously (10, 11, 40). Results of the present study is unlike that of Conlon et al. (41), which revealed that integration of IS256 to ica locus decreased the ability of strains to form biofilms. The high frequency of this transposon in this study might be due to the origin of the isolates, as we did not focus on hospital strains and some of the strains were isolated from outpatients. On the other hand, the presence of IS256 among most of the biofilm-producing strains, which were positive for icaA, icaD and atlE genes, showed the correlation between these factors and clinically relevant strains (10, 25, 42).
In conclusion, the results of this study illustrated the high prevalence of antibiotic resistant biofilm-producing S. epidermidis strains in this hospital, which could be a reservoir for antibiotic resistance genes. Furthermore, our findings suggest the importance of IS256 element as a characteristic of clinical isolates of multidrug resistant S. epidermidis, which play an important role in the adaptation of the genome in clinical isolates. On the other hand, we could not investigate the isolates relatedness in this study and further studies using genetic methods such as pulsed field gel electrophoresis (PFGE) should be done to characterize the clonal dissemination of S. epidermidis strains.