1. Background
Staphylococcus aureus is one of the most important causes of food poisoning in the world, which is due to production of a broad spectrum of staphylococcal enterotoxins (SEA-SEV) (1). Staphylococcal enterotoxins are members of staphylococcal and streptococcal exotoxins related together and consisting of pyrogenic proteins known as superantigens and may cause proliferation of the T-cells and produce cytokines. Enterotoxins are heat resistant small secretory proteins that show high resistance to digestive enzymes which enables them to be active in the gastrointestinal (GI) tract (2). Staphylococcal food poisoning is one of the most common food-borne diseases in the world characterized by vomiting, nausea, diarrhea and abdominal cramps (3, 4).
Different mobile genetic elements such as bacteriophages, plasmids and pathogenicity islands (PI) can confer the ability of production of a variety of virulence factors among S. aureus strains (1, 5). SEA, SEE, SEK, SEP and SEQ are bacteriophage encoded enterotoxins, in which SEA is the most frequent agent of food poisoning related staphylococcal enterotoxin (1). Bacteriophages convert the non-virulent bacteria to virulent ones through lysogenic phage conversion and play an important role in pathogenesis of S. aureus strains (6, 7). Staphylococcal bacteriophages belong to Myoviridae and Siphoviridae families and classification of bacteria according to the presence of different classes of prophages indicating the potential of bacteria to produce a variety of virulence factors (7, 8).
S. aureus strains can acquire resistance genes through horizontal gene transfer mechanisms which enable them to show resistance against anti-microbial agents and spread worldwide (5). Methicillin is one of the most important antibiotics, used first in 1960 to treat penicillin-resistant S. aureus strains, and first methicillin-resistant S. aureus (MRSA) strains were reported only one year after identification in 1961 and then spread in hospitals and the communities worldwide (9). Resistance to methicillin is due to the presence of mecA gene, which is a part of a large cluster called staphylococcal cassette chromosome mec (SCCmec). Eleven different SCCmec types are reported and typing of bacteria using the presence of different SCCmec types could be a useful method to classify MRSA strains (9, 10).
2. Objectives
The current experimental study aimed to isolate and evaluate the clonality of MRSA strains isolated from chicken meat samples and describe the presence of different prophage types, enterotoxin genes and also occurrence of the staphylococcus cassette chromosome mec (SCCmec) gene types.
3. Methods
3.1. Sampling and Identification of Isolates
From July to December 2014, a total of 36 chicken meat samples were collected from markets in Isfahan, Iran. All samples were collected in sterile bottles and transported to the lab in a cold chain. For sample preparation, 100 grams of each chicken meat samples were suspended in 900 mL phosphate buffered saline (PBS) and vortexed for five minutes and then filtered using 0.45 µm filter membrane (Merck Millipore, Darmstadt, Germany), as described previously (11). Filters were transported on selective chromogenic MRSA-ID agar (Oxoid Brilliance MRSA Agar, Thermo Fisher Scientific, Waltham, MA, USA) and incubated for 48 hours at 37°C. Colonies suspected of being MRSA strains were also collected and analyzed. All colonies were identified at the species level using specific ,nucA primers (12) and tested for the presence of mecA gene (13). Different primers used in this study are listed in Table 1. Moreover, all primers were synthesized by TIB Molbiol Syntheselabor GmbH (Berlin, Germany). DNA extraction of MRSA strains was carried out using a High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) according to the manufacturer instructions.
| Gene | Nucleotide Sequence (5′→3′ End) | Molecular Weight, bp | Reference |
|---|---|---|---|
| nucA | F: AGTTCAGCAAATGCATCACA | 400 | (12) |
| R: TAGCCAAGCCTTGACGAACT | |||
| mecA | F: GTAGAAATGACTGAACGTCCGATAA | 310 | (13) |
| R: CCAATTCCACATTGTTTCGGTCTAA | |||
| SCCmec type I | F: GCTTTAAAGAGTGTCGTTACAGG | 613 | (14) |
| R: GTTCTCTCATAGTATGACGTCC | |||
| SCCmec type II | F: CGTTGAAGATGATGAAGCG | 398 | |
| R: CGAAATCAATGGTTAATGGACC | |||
| SCCmec type III | F: CCATATTGTGTACGATGCG | 280 | |
| R: CCTTAGTTGTCGTAACAGATCG | |||
| SCCmec type Iva | F: GCCTTATTCGAAGAAACCG | 776 | |
| R: CTACTCTTCTGAAAAGCGTCG | |||
| SCCmec type IVb | F: TCTGGAATTACTTCAGCTGC | 493 | |
| R: AAACAATATTGCTCTCCCTC | |||
| SCCmec type IVc | F: ACAATATTTGTATTATCGGAGAGC | 200 | |
| R: TTGGTATGAGGTATTGCTGG | |||
| SCCmec type IVd | F: CTCAAAATACGGACCCCAATACA | 881 | |
| R: TGCTCCAGTAATTGCTAAAG | |||
| SCCmec type V | F: GAACATTGTTACTTAAATGAGCG | 325 | |
| R: TGAAAGTTGTACCCTTGACACC | |||
| SGA prophage type | F: TATCAGGCGAGAATTAAGGG | 744 | (8) |
| R: CTTTGACATGACATCCGCTTGAC | |||
| SGB prophage type | F: ACTTATCCAGGTGGYGTTATTG | 405 | |
| R: TGTATTTAATTTCGCCGTTAGTG | |||
| SGF prophage type | F: CGATGGACGGCTACACAGA | 155 | |
| R: TTGTTCAGAAACTTCCCAACCTG | |||
| SGFa prophage type | F: TACGGGAAAATATTCGGAAG | 548 | |
| R: ATAATCCGCACCTCATTCCT | |||
| SGFb prophage type | F: AGACACATTAAGTCGCACGATAG | 147 | |
| R: TCTTCTCTGGCACGGTCTCTT | |||
| SGL prophage type | F: GCTTAAAACAGTAACGGTGACAGTG | 748 | |
| R: TGCTACATCATCAAGAACACCTGG | |||
| sea | F: TAAGGAGGTGGTGCCTATGG | 180 | (12) |
| R: CATCGAAACCAGCCAAAGTT | |||
| Seb | F: TCGCATCAAACTGACAAACG | 478 | |
| R: GCAGGTACTCTATAAGTGCC | |||
| Sec | F: ACCAGACCCTATGCCAGATG | 371 | |
| R: TCCCATTATCAAAGTGGTTTCC | |||
| Sed | F: TCAATTCAAAAGAAATGGCTCA | 339 | |
| R: TTTTTCCGCGCTGTATTTTT | |||
| See | F: TACCAATTAACTTGTGGATAGAC | 170 | |
| R: CTCTTTGCACCTTACCGC | |||
| Seg | F: CCACCTGTTGAAGGAAGAGG | 432 | |
| R: TGCAGAACCATCAAACTCGT | |||
| she | F: TCACATCATATGCGAAAGCAG | 463 | |
| R: TCGGACAATATTTTTCTGATCTTT | |||
| Sei | F: CTCAAGGTGATATTGGTGTAGG | 529 | |
| R: CAGGCAGTCCATCTCCTGTA | |||
| Sej | F: GGTTTTCAATGTTCTGGTGGT | 306 | |
| R: AACCAACGGTTCTTTTGAGG | |||
| Sel | F: CACCAGAATCACACCGCTTA | 204 | (15) |
| R: CTGTTTGATGCTTGCCATTG | |||
| Sek | F: ATGAATCTTATGATTTAATTTCAGAATCAA | 545 | |
| R: ATTTATATCGTTTCTTTATAAGAAATATCG | |||
| Sem | F: ATGAAAAGAATACTTATCATTGTTGTTTTATTG | 720 | |
| R: CTTCAACTTTCGTCCTTATAAGATATTTC | |||
| Sen | F: ATAAAAAATATTAAAAAGCTTATGAGATTGTTC | 775 | |
| R: ACTTAATCTTTATATAAAAATACATCAATATG | |||
| Seo | F: TATGTAGTGTAAACAATGCATATGCA | 685 | |
| R: TCTATTGTTTTATTATCATTATAAATTTGCAAAT | |||
| Sep | F: TTAGACAAACCTATTATCATAATGGAAGT | 618 | |
| R: TATATAAATATATATCAATATGCATATTTTTAGACT | |||
| Seq | F: GGAAAATACACTTTATATTCACAGTTTCA | 539 | |
| R: ATTTATTCAGTTTTCTCATATGAAATCTC |
Abbreviations: SCCmec, Staphylococcus cassette chromosome mec; MRSA, methicillin-resistant Staphylococcus aureus; CT, common type.
3.2. PhP Typing
All MRSA isolates were typed using high resolution PhP system (PhPlate AB, Stockholm, Sweden) as described by Rahimi et al. (9).
3.3. SCCmec Typing of MRSA Strains
A multiplex-polymerase chain reaction (PCR) assay with eight pairs of primers (SCCmec types and subtypes I, II, III, IVa, IVb, IVc, IVd and V) (14) were used for SCCmec typing of MRSA strains according to the PCR cycle conditions introduced previously (10).
3.4. Prophage Typing of MRSA Strains
All MRSA strains were subjected to a multiplex-PCR assay with specific primers for SGA (phage type 3A), SGB (phage type 11), SGF (phage type 77), SGFa (phage type 77a), SGFb (phage type 77b), SGD (Twort-like phages) and SGL (phage type 187) prophage types as described previously (8).
3.5. Detection of Enterotoxin Genes
The presence of different enterotoxin genes (sea-seq) among MRSA strains were examined using separate PCR assays according to the protocols reported previously (12, 15).
4. Results
4.1. Identification of Isolates
In the current study, a total of 116 colonies were collected from chicken meat samples (25 samples out of 36; 69.4%). Moreover, 11 meat samples were negative for MRSA isolates. All isolates were identified as S. aureus strains using nucA specific primers and positive for the presence of mecA gene and confirmed as MRSA strains.
4.2. PhP Typing
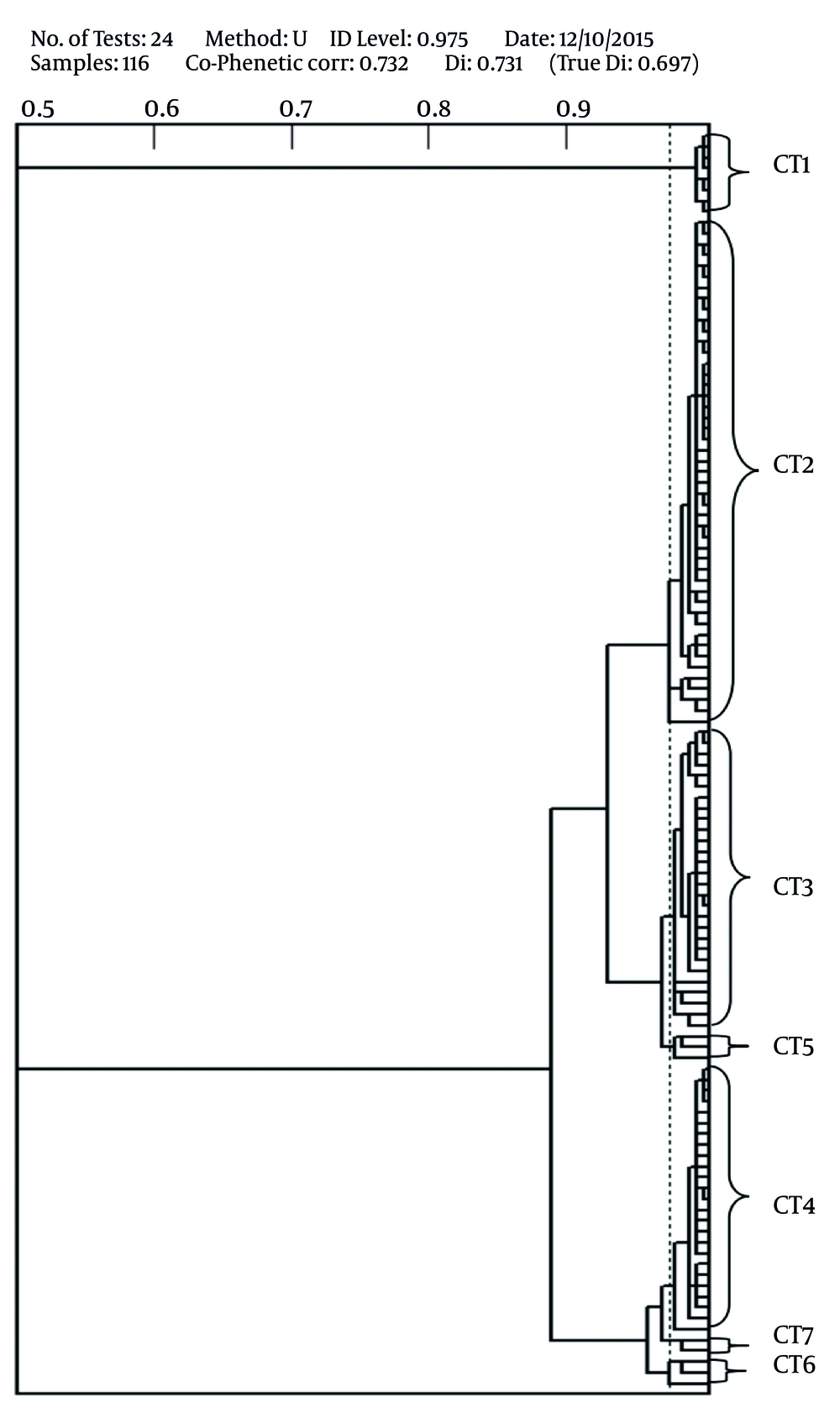
The results of PhP typing showed that all 116 MRSA isolates were discriminated into seven PhP types consisting of seven common types (CTs) and no single types (ST) were detected among the isolates (Table 2, Figure 1). Each common type consisted of 2 - 47 strains and CT2 was the dominant type in which the highest number of MRSA strains was categorized in this CT (n = 47, 41%). Twenty eight (24%) and 25 (22%) MRSA isolates were categorized in CT3 and CT4, respectively.
| PhP Types | SCCmec Type | Prophage Types | Enterotoxin Genes | Number of Isolates (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SGA | SGB | SGF | SGFa | SGFb | SGL | sea | Sek | seq | Other Genes | |||
| CT 1 | III | - | - | + | + | + | - | + | + | + | - | 8 (6.9) |
| CT 2 | III | - | - | + | + | + | - | + | + | + | - | 47 (40.5) |
| CT 3 | III | - | - | + | + | + | - | + | + | + | - | 28 (24.1) |
| CT 4 | III | - | - | + | + | + | - | + | + | + | - | 25 (21.6) |
| CT 5 | III | - | - | + | + | + | - | + | + | + | - | 3 (2.6) |
| CT 6 | III | - | - | + | + | + | - | + | + | + | - | 3 (2.6) |
| CT 7 | III | - | - | + | + | + | - | + | + | + | - | 2 (1.7) |
4.3. SCCmec Typing
In the current study, only one SCCmec type was detected among MRSA strains and all isolates harbored SCCmec type III and were classified as hospital acquired (HA)-MRSA strains (Table 2).
4.4. Prophage typing
Three different prophage types and one prophage pattern were detected among MRSA strains and 100% of the strains were positive for SGF, SGFa and SGFb prophage types (Table 2).
4.5. Detection of Enterotoxin Genes
Among all 116 MRSA strains, sea, sek and seq genes encoding SEA, SEK and SEQ were detected and all strains were negative for the presence of other eneterotoxin genes (Table 2).
5. Discussion
The current study isolated MRSA strains from chicken meat samples collected from different local markets in Isfahan in six months. The isolation of MRSA from different meat samples was reported previously from different countries (16-23). Another study (10) reported MRSA strains from poultries in Iran; therefore, the isolation of MRSA strains from chicken meat was not surprising.
Meat can become contaminated by several means reported previously. Most of the scientists indicated that meat contamination may occur during the slaughtering process of MRSA-positive animals which results in contamination of carcasses (17). On the other hand, the contamination might be due to contact of humans with large numbers of animals in the farms (24). In addition, improper handling of the meat at the slaughterhouse, processing facilities, storage of food at high temperature at the retail stores and close contact with butchers are some of the possible reasons for contamination of the meat samples (24).
All MRSA strains in the current study harbored SCCmec type III as an indicator of HA-MRSA strains. These findings were consistent with previous studies in Iran, in which SCCmec type III was the dominant type among MRSA strains isolated from clinical, environmental and animal samples (9, 10, 25-30). These findings indicate the hospital source of MRSA strains in the study.
Only one prophage pattern and three prophage types were detected in the present study and were common among all isolates. In other studies, different prophage types and prophage patterns were reported from Iran and other countries (6-9, 27, 29, 31). Although in different studies more prophage types and patterns were reported, the SGF prophage type and current prophage pattern were common among all MRSA isolates with different sources. The presence of common prophage patterns in hospitals, environments, animals and foodstuffs indicate the circulation of some clonal types in Iran. MRSA strains harboring this prophage pattern can produce different virulence factors such as enterotoxins A, K and Q, staphylokinase and beta-lysine. A previous study showed that community acquired MRSA (CA-MRSA) harbored SGA prophage type responsible for production of panton-valentine leukocidin (PVL) and this prophage type could be also another marker for CA-MRSA isolates (9).
The results of PhP typing indicated the presence of seven CTs among MRSA strains isolated from chicken meat. CT2 was the dominant PhP type (comprised of 41% of the isolates) in the present study that was in agreement with other reports from the clinics and sewages in Iran (9, 27, 28, 30) indicating an epidemiological link between the MRSA strains isolated from different sources. All seven CTs found in the present study, were also reported from clinical samples and were associated with human infections. These findings suggest that these strains were introduced to meat during meat processing or packing, subsequent to slaughtering. Different studies showed that MRSA strains are endemic in different sources in Iran; therefore contamination of meat in post-slaughtering process may not be responsible for the presence of these strains in food or animals. Other reports showed that the prevalence of STs among MRSA strains was limited to the strains that harbored SGA prophage type and also SCCmec type IV and classified as CA-MRSA strains (9, 10, 27).
Among all 116 MRSA strains, only three enterotoxin genes encoding for SEA, SEK and SEQ were detected. These enterotoxins are prophage encoded with different phage types such as ΦSa3ms, ΦSa3mw, Φ252B, ΦNM3 and ΦMu50a, which are members of SGF, SGFa and SGFb prophage types (1). It was also shown that all MRSA strains harbored SGF, SGFa and SGFb prophage types; therefore, the presence of sea, sek and seq genes was not surprising. This finding was not in agreement with those of other studies, in which the prevalence of these genes was significantly lower (12, 15, 32-41). On the other hand, the high prevalence of sea, sek and seq genes was shown in MRSA strains isolated from patients in a referral hospital in Tehran, Iran (42). The higher prevalence of sea, sek and seq genes among MRSA isolates in the current study could be, in part, due to geographical locations and different ecological settings where these studies were performed and also the fact that all 116 MRSA strains were spread from hospitals into the community and also that diversity of all strains was limited to seven CTs.
5.1. Conclusion
These findings illustrated the prevalence of clonal groups of MRSA strains in chicken meat in Isfahan, Iran. These strains were similar to MRSA strains isolated from hospitals in Iran, which indicated the important role of hospitals in dissemination of clinical isolates in the community. The presence of different prophage types among MRSA strains indicated the high potential of virulent strains to serve as reservoirs for dissemination of bacteriophage encoded enterotoxin and virulence agents.