1. Background
Klebsiella pneumoniae causes a variety of infections, in particular, the community-acquired pneumonia and nosocomial infections. Hospital-acquired infections caused by this organism include urinary tract infection, pneumonia, septicemia, and wound infection. It has been estimated that about 8% of nosocomial infections are caused by K. pneumoniae (1, 2). Given the acquisition of various resistant genes, the choice of effective antibiotics to treat infections caused by this bacterium has been limited. The broad-spectrum fluoroquinolones have been used to treat a wide range of bacterial infections (3). Fluoroquinolones bind to and inhibit the enzyme activity of topoisomerase II (DNA gyrase) and topoisomerase IV required for appropriate bacterial DNA functions. DNA gyrase is a tetramer enzyme consisting of two subunits A and two subunits B, which are coded by the gyrA and gyrB genes, respectively. Topoisomerase IV is also coded by two genes, parC and parE (4). DNA gyrase is involved in creating the negative super coiled DNA and the role of topoisomerase IV is to separate daughter chromosomes at the end of DNA replication cycles (5, 6).
Given the widespread use of fluoroquinolones, resistance to these drugs has been reported worldwide (7). So far, several mechanisms of resistance to fluoroquinolones have been identified which include mutations in topoisomerase genes (8) and reduce intracellular drug accumulation within the bacterial cell and the acquisition of plasmid-mediated quinolone-resistance genes (PMQR) (8). The mutation in topoisomerase genes is the common mechanism of resistance to fluoroquinolones and usually occurs in a certain quinolone-resistance determining region of DNA (QRDR). Mutations in gyrA are more common than gyrB gene (9). In gyrA, mutations usually occur at nucleotide 201 to 320 encoded amino acids 67 to 106, in particular at serine 83 and aspartate 87 (10). Mutations at codon 426 and 447 of gyrB usually make a low-level of resistance to fluoroquinolones (11, 12). Mutations in the amino acid 78, 80 and 84 positions of parC in K. pneumoniae resistant to fluoroquinolones have also been reported worldwide although with different rates (10, 13).
The acquisition of PMQR genes by K. Pneumoniae also plays an important role in the resistance to fluoroquinolones. Three PMQR genes have been reported which include qnr, aac(6')-Ib-cr and qepA. The qnr gene family encodes proteins that protect topoisomerase II and IV from quinolones impacts. The aac(6')-Ib-cr gene, which was first discovered in 2003, decreases the activity of fluoroquinolones including ciprofloxacin and norfloxacin (14, 15). The qepA gene causes a high level of resistance to hydrophilic fluoroquinolones (16).
2. Objectives
To better understand the role of these mechanisms in antibiotic resistance, this study aimed to determine mutations in the QRDR of gyrA, gyrB and parC genes and the frequency of PMQR genes in K. pneumoniae isolates resistant to fluoroquinolones.
3. Methods
3.1. Bacterial Isolates
In this experimental study, One hundred non-duplicate isolates of K. pneumoniae were collected from 2013 to 2014 in three general hospitals in Kermanshah during an increase of K. pneumoniae infection among hospitalized patients. The clinical samples included urine (n = 54, 54%), burn (n = 15, 15%), respiratory tract secretions (n = 15, 15%), and others (blood, wound and ascitic fluid) (n = 16, 16%). They were collected form 59 (59%) female and 41 (41%) male people with the average age of 39.5 ± 26 years old. All isolates were identified by bacteriological and biochemical tests followed by confirmation using API20E Kit (bio-Merieux, France).
3.2. Antibiotic Susceptibility Testing
Minimum inhibitory concentrations (MIC) of levofloxacin and ciprofloxacin (Sigma, USA) were determined by a standard broth microdilution method according to CLSI protocols (17). Briefly, Mueller-Hinton broth (Merck, Germany) with a final inoculum of 5 × 105 CFU/mL in 96-well plates were incubated at 37°C for 24 hours. E. coli strain ATCC 25922 was used as the standard drug- susceptible control for MIC measurements. The breakpoints recommended by CLSI were used for ciprofloxacin (susceptible ≤ 1 µg/mL; resistant ≥ 4 mg/mL) and levofloxacin (susceptible ≤ 2 µg/ mL; resistant ≥ 8 mg/mL).
3.3. DNA Extraction and PCR Assay
The DNA of isolates resistant to flouroquinolones was extracted using the boiling method. Amplification of the QRDR of gyrA, gyrB, and parC genes was carried out by PCR using specific primers (SinaClon, Iran) (Table 1). The PMQR genes (qnrA, qnrB, aac(6´)-Ib and qepA) were amplified using specific primers by PCR (SinaClon, Iran) (Table 1). After electrophoresis of PCR products on 1% agarose gel (Merck Co, Germany) and staining with ethidium bromide, the gels were visualized by gel-Documentation apparatus (BioRad, USA). PCR products were sequenced and analyzed by BLAST tools (http:// blast.ncbi.nlm.nih.gov/Blast.cgi).
| Primer | DNA Sequence 5′-3′ | Target Site | Amplicon Size, bp | Reference |
|---|---|---|---|---|
| gyrA6 | CGA CCT TGC GAG AGA AAT | QRDR of gyrA | 626 | (18) |
| gyrA631R | GTT CCA TCA GCC CTT CAA | |||
| gyrB-F | TCTACTGCTTYACCAACAACA | QRDR of gyrB | 704 | (19) |
| gyrB-R | CGTCCGCATCGGTCATGAT | |||
| parCF | ATGGCAGAGCGCCTTGCG | QRDR of parC | 437 | (19) |
| parCR | GCCGTCRAAGTTTGGCAC | |||
| qnrA-F | ATTTCTCACGCCAGGATTTG | qnrA | 516 | (20) |
| qnrA-R | GATCGGCAAAGGTTAGGTCA | |||
| qnrB-F | GATCGTGAAAGCCAGAAAGG | qnrB | 469 | (20) |
| qnrB-R | ACGATGCCTGGTAGTTGTCC | |||
| qnrS-F | ACGACATTCGTCAACTGCAA | qnrS | 417 | (20) |
| qnrS-R | TAAATTGGCACCCTGTAGGC | |||
| aac(6′)-Ib-F | TTGCGATGCTCTATGAGTGGCTA | aac(6′)-Ib | 482 | (15) |
| aac(6′)-Ib-R | CTCGAATGCCTGGCGTGTTT | |||
| qepA-F | GGACATCTACGGCTTCTTCG | qepA | 720 | (14) |
| qepA-R | AGCTGCAGGTACTGCGTCAT |
Abbreviations: bp, base pair; F, forward primer; R, reverse primer.
3.4. Restriction Digestion
RFLP was used to differentiate the naive aac(6´)-Ib gene from those of the cr variant of this gene involved in flouroquinolones resistance. All PCR products for aac(6´)-Ib gene were digested by BtsCI restriction endonuclease (Fermentase, England). The presence of two DNA fragments with 272 and 210 bp by digestion indicated of aac(6´)-Ib, whereas an undigested DNA indicated of aac(6´)-Ib-cr variant.
3.5. Sequencing and Data Analysis
All PCR products for gyrA, gyrB, and parC genes were purified using a DNA purification kit and sequenced (SinaClon, Iran). The DNA sequence was performed using an ABI 3730XL DNA analyzer (Macrogen Inc, Korea). Sequence data were analyzed for homology using the national center for biotechnology information GenBank database (http://www.ncbi.nlm.nih.gov/). The nucleotide sequence data have been deposited in the pubmed genebank nucleotide sequence database (http://www.ncbi.nlm.nih.gov/).
All data were statistically analyzed using SPSS version 20 for associations between mutations with the PMQR genes and the level of flouroqinolone resistance by Kruskal-Wallis and Mann-Whitney tests.
4. Results
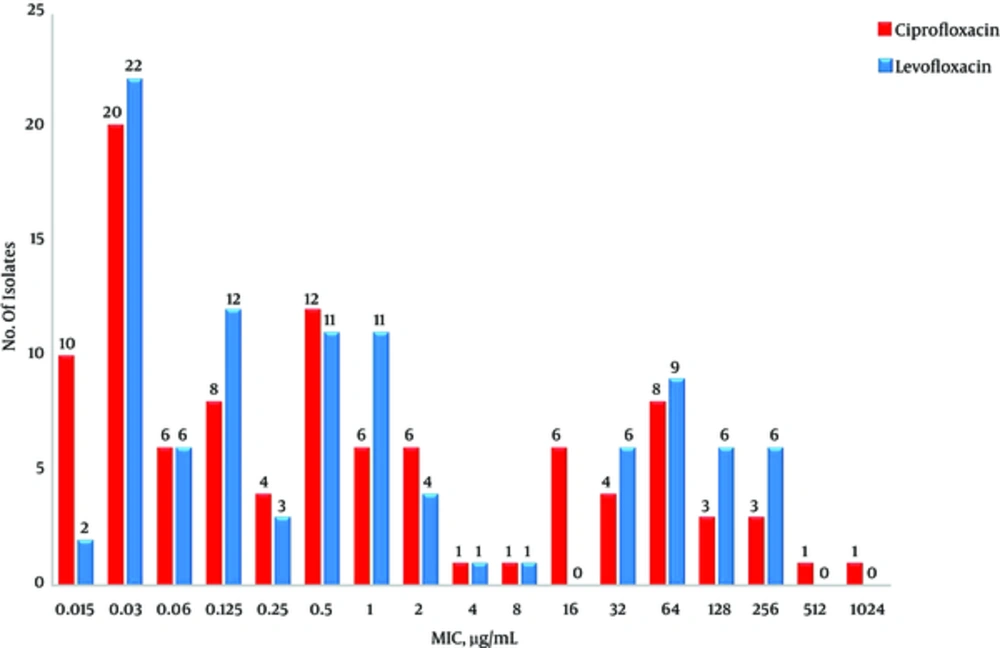
The antibiotic susceptibility and MIC of ciprofloxacin and levofloxacin for 100 K. pneumoniae isolates have been shown in Table 2 and Figure 1.
aThe MIC at which 50% of the isolates are inhibited.
bThe MIC at which 90% of the isolates are inhibited.
DNA sequence analysis of the QRDR of gyrA showed mutations at codon 83 of 24 (85.7%) and at codon 87 of 20 (71.4%) isolates (Table 3). Sequence analysis of parC showed 15(53.6%) isolates with a mutation at codon 80 and 7 (25%) isolates with a mutation at codon 84.
| Gene | Amino Acid Position | Nucleotide Changes | Amino Acids Substitute | No. of Isolates (%) |
|---|---|---|---|---|
| gyrA | Serine 83/ | TCC → TTC | Phenylalanine | 12 (42.9) |
| Asp87 | GAC → GCC | Alanine | ||
| Serine 83/ | TCC → TTC | Phenylalanine | 1 (3.6) | |
| Asp87/ | GAC → GCC | Alanine | ||
| Glu94 | CAG → CAC | Histidine | ||
| Serine 83/ | TCC → TTC | Phenylalanine | 4 (14.3) | |
| Asp87/ | GAC → AAC | Asparagine | ||
| Serine 83/ | TCC → TTC | Phenylalanine | 3 (10.7) | |
| Asp87/ | GAC → AAC | Asparagine | ||
| Thr161 | ACC → ATC | Isoleucine | ||
| Serine 83 | TCC → ATC | Isoleucine | 2 (7.1) | |
| Serine 83 | TCC → TAC | Tyrosine | 2 (7.1) | |
| Glu94 | CAG → CAC | Histidine | 1 (3.6) | |
| WT | - | - | 3 (10.7) | |
| gyrB | Asp368 | GAC → AAT | Asparagine | 1 (3.6) |
| WT | - | - | 27 (96.4) | |
| ParC | Ser80 | AGC → ATC | Isoleucine | 15 (53.6) |
| Glu84 | GAA → AAA | Lysine | 7 (25) | |
| WT | - | - | 6 (21.4) |
zAbbreviation: WT, wild type.
In three isolates, an extra mutation outside QRDR in gyrA at position 161 was detected which did not make significant changes in MIC of ciprofloxacin and levofloxacin in comparison with isolates without this mutation (P = 0.645). Similarly, one isolate with an extra mutation outside QRDR in gyrA at codon 94 had no significant impact on MIC compared with the isolates without this mutation for both antibiotics tested (P = 0.610). Sequence analysis of gyrB showed a mutation outside QRDR in one isolates at codon 368, which apparently did not increase bacterial MIC.
The qnrB and qnrS were found in 11 (39.3%) and 1 (3.6%) isolates, respectively. Of the 28 isolates selected for detection of the aac(6')-Ib-cr, 25 of 28 (89.3%) isolates were positive for aac (6')-Ib and all of them was the aac(6')-Ib-cr variant. PMQR genes enhanced the resistance of isolates to fluoroquinolones (Table 4).
| PMQR Genes | Mutations in gyrA and/or parC | MIC, μg/mL Range | No. of Isolates (%) | |||||
|---|---|---|---|---|---|---|---|---|
| CIP | LVX | |||||||
| 4 - 16 | 32 - 128 | > 256 | 4 - 16 | 32 - 128 | > 256 | |||
| qnrB+ | Ser83 + Asp87 + ser80 + Asp368 | 1 | 1 | 1 (3.6) | ||||
| aac-Ib-cr | ||||||||
| qnrB+ | Ser83 + Asp87 + ser80/Glu84 | 4 | 2 | 4 | 2 | 6 (21.4) | ||
| aac(6´)-Ib-cr | ||||||||
| aac(6´)-Ib-cr | Ser83 + Asp87 + ser80/Glu84 | 3 | 3 | 1 | 5 | 2 | 7 (25) | |
| - | Ser83 + Asp87 + ser80/Glu84 | 1 | 1 | 2 | 2 (7.1) | |||
| aac(6´)-Ib-cr | Ser83 + Asp87 + Thr161 + Glu84 | 3 | 1 | 3 (10.7) | ||||
| aac(6´)-Ib-cr | Ser83 + Asp87 + Glu94 + ser80 | 1 | 1 | 1 (3.6) | ||||
| qnrB+ | Ser83 + ser80 | 1 | 1 | 1 (3.6) | ||||
| aac(6´)-Ib-cr | ||||||||
| - | Ser83 + ser80 | 1 | 1 | 1 (3.6) | ||||
| qnrB+ | Ser83 | 1 | 1 | 1 (3.6) | ||||
| aac(6´)-Ib-cr | ||||||||
| aac(6´)-Ib-cr | Ser83 | 1 | 1 | 1 (3.6) | ||||
| aac(6´)-Ib-cr+ | - | 1 | 1 | 1 (3.6) | ||||
| qnrS | ||||||||
| qnrB+ | - | 1 | 1 | 1 (3.6) | ||||
| aac(6´)-Ib-cr | ||||||||
| aac(6´)-Ib-cr | - | 1 | 1 | 1 (3.6) | ||||
| qnrB+ | Glu94 | 1 | 1 | 1 (3.6) | ||||
| aac(6´)-Ib-cr | ||||||||
5. Discussion
The results of previous studies indicated that the rate of resistance to ciprofloxacin is higher than levofloxacin among Enterobacteriaceae (21, 22). However, in our study, all isolates resistant to ciprofloxacin were also resistant to levofloxacin but the average MIC for ciprofloxacin was higher than levofloxacin that indicate levofloxacin is slightly more effective against K. pneumoniae isolates.
The impact of mutations in DNA gyrase and topoisomerase IV on the resistance to fluoroquinolones in bacteria have previously been studied (23). In Gram-negative bacteria, mutations usually occur in subunit GyrA protein of DNA gyrase which is the first target of fluoroquinolones. DNA sequence analysis of our data showed 5 types of mutations in gyrA gene with amino acid changes among K. pneumoniae isolates which is consistent with the results of previous studies (13, 19). Mutation at codon 83 in gyrA was the most frequent one among fluoroquinolone resistant K. pneumonia and substitution of the serine by phenylalanine was the most common changes. This result is consistent with the results of other studies (24). Moreover, mutation outside QRDR of gyrA was also detected in three strains at codon 161. Another Iranian paper has reported this mutation among K. pneumoniae isolates (24).
The MIC of fluoroquinolones for isolates with two mutations in gyrA gene was higher than isolates with only one mutation that is similar to other studies’ results (25-28). Sequence analysis of QRDR in parC gene revealed two mutations with amino acid changes which have been reported by several previous studies (19, 29). In the present study, all isolates with mutations in parC showed also mutations in gyrA that may suggest the topoisomerase IV is the second target for the fluoroquinolones (12). The isolates with mutations in both gyrA and parC simultaneously showed the higher MIC levels, which is compatible with the results of previous studies (13, 30). In three isolates, mutations in gyrA outside of QRDR were detected, but these mutations apparently do not significantly change the MIC value in comparison with isolates without mutations. In three isolates, MIC level was high, but without any mutations in the QRDR of gyrA, gyrB and parC genes. Since the plasmid genes can cause a low level of resistance, there may be other resistance mechanisms that involve such efflux pumps and mutation in parE gene as it has been reported by other studies. For instance, Al-Marzooq et al. and Park et al. have reported the K. pneumoniae isolates with resistant to ciprofloxacin but with no mutations in gyrA and parC. They suggested PMQR genes play an important role in the resistance to ciprofloxacin (13, 31).
The aac (6') -Ib-cr gene has recently been reported among bacteria and was the most common gene in our study. This is consistent with the results of other studies in Iran, South Korea and Sweden (32-34). In some studies in China and Thailand a low prevalence for aac (6') -Ib-cr has been reported (35, 36). Similarly, in these studies the frequency of aac (6') -Ib-cr was also higher than other genes. The frequency of qnrB gene in our study was high that is supported by the results of other studies (37, 38). On the other hand, the qnrS gene had a very low frequency, which is also similar to the results of previous studies, including studies in Iran, South Korea and China (33, 35). Furthermore, qnrA and qepA genes has not been found in several studies, including in Iran, Korea, Malaysia, which support our data for these genes (38, 39).
5.1. Conclusions
In conclusion, resistance to fluoroquinolones is relatively high among K. pneumoniae in Kermanshah. The MIC of isolates for fluoroquinolones increases by the accumulation of mutations within QRDR and harboring more PMQR genes. It seems that mutations in QRDR of topoisomerase genes play an important role in K. pneumoniae resistant to fluoroquinolones. The plasmid mediated genes also increase the level of resistance. The mutations outside the QRDR of topoisomerase genes also occur in K. pneumoniae isolates but their exact role in resistance is not clear.