1. Background
Inflammatory bowel disease (IBD) includes various forms of disorders; however crohn’s disease (CD) and ulcerative colitis (UC) are 2 major types of this disease (1). There are some evidences regarding the role of genetic and immune disorders and environmental factors, such as change in fecal microbiota, in the pathogenesis of IBD (2). The humans gut microbiota comprises different bacterial populations that live in a peaceful coexistence (3). This is estimated that the adults’ human gut contains around 1014 bacterial cells and over 1000 different bacterial species (4, 5). Colonization of the gut microbiota begins after birth in babies, is stabilized during the first years of life, and remains stable throughout our life (6). Role of the gut microbiota in the occurrence of IBD has yet to be determined. The commensal microbiota produces antigens, designated as pathogen associated molecular patterns (PAMP), which stimulate an immune system that subsequently causes chronic intestinal inflammation (7, 8). In this subject, dominance of some bacterial phyla could stimulate a more severe inflammatory response than the others. Firmicutes and Bacteroidetes are the most dominate bacterial phyla in the colon (9). Changes of the quantity of commensal bacteria in IBD patients, especially within members of the Firmicutes, Bacteroidetes, and Proteobacteria phyla, were reported in several studies (10-13). Alterations in the abundance of some specific phyla, e.g. reduction of Bacteroidetes and Firmicutes and increase in Gammaproteobacteria, were reported in patients with CD or UC (14). However, newer studies at genus levels revealed some diversity among members of these phyla.
Streptococcus is a member of Gram-positive lactic acid-producing bacteria (LAB) that belonged to Firmicutes phylum. Many strains of Streptococcus are non-pathogenic and occur as commensal flora on the skin, the oral cavity, nasopharynx, upper respiratory tract, urogenital, and gastrointestinal tracts. Some species of Streptococcus are responsible for numerous human inflammatory diseases including pharyngitis, meningitis, endocarditis, necrotyzing fasciitis, and those that are immune mediated, such as acute glumeronephritis, acute rheumatic fever, and colon cancer (15, 16). Interaction of some virulence factors of Streptococci with immune cells could elicit inflammatory response in different organs (17, 18). Association of Streptococcusbovis with IBD and colon cancer was reported in previous studies (19). In one study, it was shown that Streptococci are the predominant group of bacteria in inflamed colonic mucosa of CD patients; however this association was not verified in patients with UC (20). Contrary to these results reduction of Streptococci in IBD patients were observed in studies that investigated health-beneficial effects of these bacteria (20). Streptococci may promote this involvement through modulation of secretion levels of inflammatory cytokine in the mucosa of gastrointestinal tract (GIT). In this study, to determine involvement of Streptococcus spp. in the occurrence of IBD, prevalence and relative abundance of Streptococcus spp. were determined in fecal samples of IBD patients and healthy volunteers using relative quantification method.
2. Methods
2.1. Sampling and Processing
To investigate diversity of Streptococci, freshly prepared fecal samples of 29 IBD patients and 29 healthy volunteers were obtained from IBD patients and healthy volunteers in Taleghani hospital, Tehran, Iran in 2015. All IBD patients were characterized by both colonoscopic and pathologic criteria in the gastroenterology unit. To measure disease activity of UC and CD, criteria that were proposed by Sutherland and WR. Best et al. were used, respectively (21, 22). Demographic data were collected by a questionnaire to provide data about underlying diseases, medication and antibiotic usage, surgery of GIT, gender, and age. All samples were collected in a clean container and transferred to the laboratory during 1 hour. Total DNA was extracted from the fecal samples using DNA extraction kit (Bioneer, South Korea) and the DNA samples were stored at -20°C until use for further investigations.
This study was approved in research center for gastroenterology and liver disease Shaheed Beheshti University of Medical Sciences with ethical consideration code 841.
2.2. Relative Abundance of Streptococcus spp. by Real-Time Quantitative Polymerase Chain Reaction
Primer set suitable for real time PCR was selected for Streptococcus spp. to compare the abundance of this genus of bacteria in IBD patients and healthy controls. The oligonucleotide primers used in this study were Streptococcus spp. forward primer 5’-AGATGGACCTGCGTTGT-3’ and reverse primer 5’-TGCCTCCCGTAGGAGTCT-3’, which target 130 bp of 16s rRNA gene in most species of Streptococci (23) and Enterococcus spp. forward primer 5’-CCCTTATTGTTAGTTGCCATCATT-3’ and reverse primer 5’-ACTCGTTGTACTTCCCATTGT-3’ was selected as endogenous control (24).
Total concentration of DNA extracts were adjusted before performance of the assay. To prevent formation of primer dimer and non-specific products during real time PCR, melt curve analysis and PCR were done. Amplification of target sequences for Streptococci and endogenous control (144 bp of 16SrDNA of Enterococcus genus) was performed in separate reactions in each 96-well plate read using Applied Biosystems 7500 Real-Time PCR System. An equal amount of E. faecalis DNA (ATCC 512999) was added to the adjusted DNA samples to overcome normalization of data by ABI 7500 software on the basis of its diversity among the samples. DNA extract of a randomly selected control sample was used as a calibrator (reference sample) in all the experiments. Two separate replica of the assay were performed for all the experiments. To enable relative quantitation (RQ) analysis, average amounts of Streptococcus spp. threshold cycle (CT) in each sample were compared with CT of Streptococci in the calibrator sample. Furthermore, a cut off value was calculated based on mean RQ ± 2 SD of Streptococcus spp. in healthy controls to estimate overgrowth of this bacterium in the IBD patients.
2.3. PCR and Real Time PCR Conditions for Streptococci and Enterococcus spp.
To evaluate quality of the primers for identification of target genes in each bacterial genus, PCR was done. The 25 μL PCR reaction mixture contained 2.5 μL of 10X PCR buffer, 1.25 μL of each primer (10 picomol), 0.5 μL of each deoxyribonucleotide triphosphate (dNTP, 10 mM), 0.75 μL MgCl2 (50 mM), 0.75 U of TaqDNA polymerase (5 U/mL), and 1 μL of DNA template from pure cultures of the reference strains. It was performed in a thermal cycler (AG 22331; Eppendorf, Hamburg, Germany). A reaction with no template DNA was included as control (NTC) reaction. The PCR conditions was as follow: 1 cycle of initial denaturation at 93°C for 1 minutes, 35 cycles of denaturation at 95°C for 45 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 35 seconds. Final extension was performed at 72°C for 10 minutes. PCR products were analyzed by electrophoresis in a 1.2% agarose gel after staining using ethidium bromide (0.5 μg/mL). Differences of the primers in amplification were calculated by band volume analysis using UVIgeltec software (version 12.1).
The real time PCR for relative quantification of target bacterial genus was performed in a total volume of 20 μL using 10 µL SYBR® Premix Ex TaqTM master mix (TaKaRa, Japan), 0.4 µL of each forward and reverse primer, 0.08 µL diluted ROX, and 2 µL of DNA template under following conditions: 1 step of initial denaturation at 95°C for 30 seconds, and 35 cycle repeats of denaturation at 95°C for 5 seconds, annealing at 62°C for 34 seconds, and extension at 60°C for 34 seconds. The reaction was ended after final extension at 75°C for 15 seconds. To analyze formation of primer dimmer or non-specific products during the amplification, melt curve analysis was performed. Accordingly, at the end step of the amplification, the reaction was continued by heating at 95°C for 15 second, a temperature gradient of 60 -95°C for 1 minutes, and 95°C for 15 second.
2.4. Statistical Analysis
Relative abundance of the bacteria in the control and patient groups was analyzed by the ABI 7500 software. Statistical difference between these groups was determined by the GraphPad prism 5 using an unpaired t-test. Receiver-operator characteristic (ROC) curve was drawn to show sensitivity and specificity of the test for differentiation of IBD patients from control people.
3. Results
3.1. Patients and Sampling
A total of 29 fecal samples from IBD patients (22 with UC and 7 with CD) and 29 healthy volunteers, which show no history of other gastrointestinal disorders, were collected in this study. Among the patients with IBD, 13 patients were in the clinical remission and 16 in the flare up stages. In the IBD group, the median age was 35 years (range, 22 to 61 years), while mean age of 31 years old was measured in the control group. Association between the patients’ demographic data and disease severity was shown in Table 1.
| Variables | Crohn’s Disease (N = 7) (Remission) (Flare up) | Ulcerative Colitis (N = 22) (Remission) (Flare up) | Control | P Valuea | ||
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 1/3 | 2/3 | 3/10 | 7/10 | 20 | |
| Female | 3/4 | 1/4 | 6/12 | 6/12 | 9 | |
| Age, y | ||||||
| 10 - 20 | - | - | - | - | - | |
| 20 - 40 | 3/5 | 2/5 | 6/16 | 10/16 | 15 | |
| 41 - 60 | 1/2 | 1/2 | 3/6 | 3/6 | 14 | |
| > 60 | - | - | - | - | 2 | |
| BMI | 0.02 | |||||
| Overweight | 1/3 | 2/3 | 3/6 | 3/6 | 6 | |
| Obesity | 1/1 | - | 1/4 | 3/4 | 2 | |
| Normal | 1/1 | - | 5/11 | 6/11 | 21 | |
| Underweight | 1/2 | 1/2 | - | 1/1 | ||
| Disease duration | ||||||
| ≤ 6, mo | - | - | - | 1/1 | ||
| 7 - 12, mo | - | 1/1 | 1/4 | 3/4 | ||
| 1 - 5, y | 1/1 | - | 4/9 | 5/9 | ||
| > 5, y | 3/5 | 2/5 | 4/8 | 4/8 | ||
| Familial history | 1/1 | - | 1/1 | - | ||
| Activity indexb | 0.004 | |||||
| 0 - 2 | NA | NA | 8/9 | 1/9 | ||
| 4 - > 12 | NA | NA | 1/12 | 11/12 | ||
| Polyp | - | - | - | - | ||
| Atrophy | 1/1 | - | 1/1 | - | ||
| Dysplasia | - | - | - | - | ||
| Abscess | 2/2 | - | 5/10 | 5/10 | ||
| Surgery of GIT | 2/4 | 2/3 | 2/9 | 5/13 | ||
Abbreviations: BMI, Body Mass Index; GIT, Gastrointestinal Tract.
aP value ≤ 0.05 was shown to present association of demographic data and IBD.
bDisease activity was measured based on criteria were proposed by Sutherland and WR. Best et al..
3.2. Characterization of Streptococcus spp.
Results of conventional PCR showed usefulness of the primers for detection of Streptococcus and Enterococcus spp. both in the pure cultures of reference strains and in DNA extracts of the faecal samples (supplementary file appendix 1). Melt curve analysis results refused formation of any non-specific amplicon or primer dimer during the assay (supplementary file appendix 2). Analysis of the Ct value for Enterococci (endogenous target gene) didn’t show significant changes among the samples (supplementary file appendix 3).
PCR product of Streptococcus spp. was sequenced and number KJ6661232.1 was obtained after submission to gene bank database.
3.3. Relative Abundance of Streptococcus spp. in IBD Patients and Healthy Controls
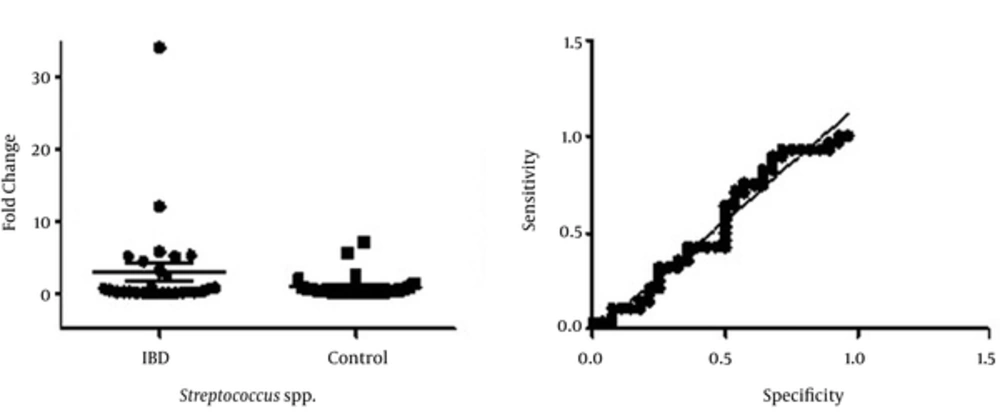
As was depicted in Figure 1, relative increase in abundance of Streptococcus spp. is seen among IBD patients (0.00064 to 34.219 fold increase) compared with controls (ranged between 0.0055 to 2.499 fold). Ct cutoff were selected on cycle 30 for exclusion of non-specific amplicons. Considering the calculated cut off value of 1.07 ± 0.31, overgrowth of Streptococcus spp. was detected in 27% of the patients with UC (6/22) and 42% in those with CD (3/7). Overgrowth of Streptococcus spp. was detected in (5/29) 17% of the control groups. There was no statistically significant difference determined between the IBD patients and samples of the control group (P value 0.1).
3.4. Relationship Between Activity Index Score, Body Mass Index, and Age
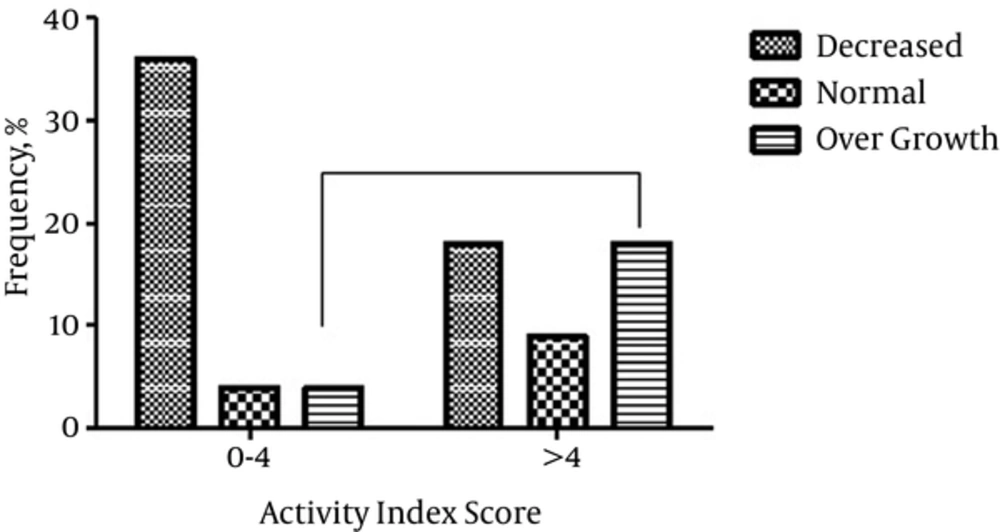
Among UC patients that have activity index score > 4, overgrowth of Streptococci was detected among 4 patients (18%), while normal abundance or decreased level of this bacterium were observed among 2 (9%) and 4 (18%) patients, respectively. While increased amount of Streptococci was shown, nearly 4.4 fold higher in patients with disease at active phase (activity index > 4), no significant association was found between the increased abundance and severity of UC based on this score (Figure 2). Analysis of body mass index (BMI) indicated that there was significant difference among the cases from IBD and normal groups (P value 0.02). This difference was not associated with overgrowth of Streptococci in the UC and CD patients (Table 1, P value 0.4). Diversity of BMI in each group was shown in supplementary file appendix 4. Changes in abundance of Streptococci were not also varied significantly among distinct age groups (supplementary file appendix 5).
Usage of new technology is needed to characterize, identify, and qualify association between different variables in the occurrence inflammatory bowel disease. These methods are mainly culture independent, which are not as firmly known as gold standard method for conventional microbial study.
4. Discussion
In the present study we compared relative abundance of Streptococcus spp. in patients with IBD and in healthy individuals using Real-Time PCR. The results determined a relative increase in Streptococcus spp. population in IBD patients that is not related to other elements that may affect disease such as BMI in IBD patients or disease activity. This finding supports evidence of association between gut microbiota and IBD.
Diversity of colonic mucosa associated bacterial microflora in Crohn’s disease was shown by Ott SJ et al. using 16S rDNA based single strand conformation polymorphism (SSCP) fingerprint, cloning experiments, and real time PCR (25). Intestinal dysbiosis, i.e. abnormal balance of common microbiota in the intestine, consisting of a decrease in beneficial bacteria such as Bifidobacterium population and their metabolic end-products, together with an increase of pathogenic bacterial populations and their toxic metabolites was reported by several studies in IBD patients (26-29). While there are several reports regarding increased levels of Gram negative bacteria, such as members of Enterobacteriacae and Proteobacteria, including adherent and invasive E. coli, Pseudomonas spp., and Yersinia, in IBD patients, few Gram positive bacteria, e.g. Mycobacterium avium subspecies paratuberculosis (MAP), have shown that their increase are implicated in the pathogenesis of IBD (30). In a study by Seungha Kang et al. using microarray analyses, decreased abundance of some bacteria, including some species of Clostridia, Eubacterium rectale, Ruminococcus albus, R. callidus, R. bromii, Faecalibacterium prausnitzii, as well as Bacteroidetes, and increased frequency of some Gram positive bacteria, including Enterococcus spp., Listeria, C. difficile, and Lactobacillus was determined in Crohn’s disease patients (31). These results showed the importance of both of the 2 bacterial groups in occurrence or progress of IBD.
Streptococci compose most predominant bacteria of oral cavity and one of prevalent bacteria in the human GIT; however few data exist on their abundance and roles in GIT of IBD patients. Pro-inflammatory responses are a frequent outcome of Streptococcal interactions with a range of host cells. Some species of this bacterium, e.g. Streptococcus suis (S. suis), showed association with IBD (32). Recently, it was shown that subtilisin-like protease (SspA) of S. suis can induce pro-inflammatory response in macrophage through Toll-like receptor 2 (TLR2) (33). Sybille Landwehr-Kenzel et al. determined a higher expression level of TLR2 in the intestine of IBD patients compared with healthy people (34). This finding could indirectly explain the higher score of activity index among our patients through interaction of TLRs with elevated amounts of Streptococci in their intestine. M protein of Streptococcus group A, rhamnose glucose polymers, glucosyltranferase, and AgI/II polypeptides of S. mutans, are other components of Streptococci that are able to promote the inflammatory response in human tissue. Streptococcus agalactiae, Group B Streptococcus, is another member of these bacteria that present as commensal bacteria of intestinal tracts in 15% - 30% of healthy adults (34). Lipoprotein and other components of this bacterium are able to induce pro-inflammatory response in the intestine. In IBD patients, direct or indirect interaction of Streptococcus metabolites and structural components at higher amounts with different cellular counterparts, which are involved in inflammatory response, including NOD, nucleotide oligomerization domains, and TLRs, which could promote early onset disease. Further studies are needed to establish the association of intestinal colonization of Streptococci with occurrence of IBD and inflammatory response in these patients.
Association between BMI and IBD was established in several studies (35-38). Similar to our results, while direct association between increased BMI and IBD was shown in most of these investigations, decreased BMI was reported among UC patients in a meta-analysis study by Jie Dong et al. (39). In our study, increased BMI was detected between both CD and UC patients, which was nearly 2 folds higher than those detected in control samples. This association was not related to the increased level of Streptococcus spp. Involvement of some bacterial genera other than Streptococci, including Eubacterium ventriosum et rel., Roseburia intestinalis et rel., and Clostridium cluster IV seems to be probable in this subject (40). More studies are needed to explain the association of changes of gut microbiota with increased BMI in IBD patients.
Collectively, results of our study demonstrated a relative increase in abundance of Streptococci in IBD patients compared with healthy volunteers. This increase was correlated with a higher activity index in UC patients. A direct association was determined between IBD and elevated BMI, which was not correlated to the increased amounts of Streptococci. Further studies are needed to reveal involvement of Streptoccous spp. virulence factors in occurrence and severity of IBD complications in different populations.
4.1. Conclusion
The verified association between increases in abundance of Streptococcus spp. and IBD suggests possible involvement of this bacterial genus in promotion or severity of the disease