1. Background
Acinetobacter baumannii has become an important nosocomial pathogen responsible for hospital outbreaks during the past few decades. In fact, A. baumannii is the fifth most common pathogen which targets the critically ill patients in intensive care units (ICU) worldwide (1, 2). Multidrug-resistant strains of A. baumannii possessing a variety of resistance mechanisms to all existing antibiotic classes including aminoglycosides, quinolones, and broad-spectrum β-lactams, have become a frequent problem in clinical settings (3, 4). In addition, due to its innate resistance to desiccation and disinfectants, A. baumannii has the ability to survive in the environment for extended periods and is almost impossible to eradicate from the hospital settings (5). The outstanding ability of A. baumannii to acquire a great variety of resistance mechanisms including chromosomal mutations or acquisition of genetic elements such as plasmids, transposons or integrons, contributes to the emergence of multidrug-resistant strains.Integrons are genetic elements that recognize and capture mobile gene cassettes, including the antimicrobial drug resistance determinants (6). Among the antibiotic-resistant integrons, classes 1 and 2 are most frequently associated with multidrug-resistant pathogens. These integrons are often located on plasmids that can facilitate their horizontal transfer among bacteria (7). Class 1 integrons are commonly found in A. baumannii and typically encode genes for aminoglycoside resistance, Ambler class A β-lactamases, metallo-beta-lactamases, and oxacillinases as well resistance to antiseptics and sulfonamides (8-11). Class 2 integrons, embedded within the transposon Tn7, display a narrower diversity of gene cassettes; and those described in A. baumannii have shown to confer resistance to aminoglycosides, chloramphenicol, trimethoprim, streptothricin, and streptomycin (12).
2. Objectives
The aim of this research was to study the correlation between plasmid carriage and presence of class 1 and 2 integrons in nosocomial isolates of A. baumannii.
3. Methods
3.1. Bacterial Isolates
Fifty isolates of A. baumannii were collected from Imam Hossein hospital in Tehran between October 2014 and March 2015. The majority of the isolates were obtained from the intensive care unit (ICU, n = 45) and were mostly from sputum (n = 34) followed by blood (n = 6), catheters (n = 3), trachea (n = 3), urine (n = 2), and wounds (n = 2). Thirty eight isolates (76%) were from men and 12 (24%) were from women. Bacterial identification was carried out by standard biochemical tests as well as detection of the blaOXA-51 gene (13). The isolates were stored at -20°C in brain heart infusion broth containing 8% dimethyl sulfoxide (v/v) until use.
3.2. Molecular Identification of A. baumannii
Detection of blaOXA-51 gene among the isolates was carried out using primers listed in Table 1 (14). DNA extraction was carried out by boiling (15). Briefly, a loopful of an overnight grown culture of each test isolate was suspended in 500 µL of sterile double-distilled water, boiled at 100°C for 10 minutes, and centrifuged at 13,000 × g for 10 minutes. The supernatant was then used as the DNA template for PCR amplification. PCR reaction mixtures (25 µL) contained 1 µL of DNA template, 1.4 mM MgCl2, 0.24 mM of each dNTP, 4 pM primer, and 2.5 U of Taq DNA polymerase in the buffer provided by the manufacturer (CinnaGen, Iran). The amplifications were performed in a thermocycler (Techne TC-3000G; UK) with the following program: initial denaturation at 94°C for 5 minutes, 30 cycles at 94°C for 25 seconds, 52°C for 40 seconds, and 72°C for 50 seconds, followed by a 6 minutes final elongation step at 72°C. PCR products were separated on 1% agarose gels and visualized after staining with RedSafe (iNtRON Biotechnology, Korea) using an image analysis system (UVLtec; St John’s Innovation Centre, UK).
3.3. Antibacterial Susceptibility
Disc diffusion was employed to determine the susceptibility of the isolates to 18 antibiotics as recommended by the CLSI 2015 guidelines (17). The antibiotic discs were: azteronam (ATM, 30 µg), amoxicillin/clavulanic acid (AUG, 30 µg), ceftazidime (CAZ, 30 µg), cefotaxime (CTX, 30 µg), cefepime (CPM, 30 µg), carbenicilin (PY, 100 µg), ampicillin/sulbactam (SAM, 10 µg), piperacillin (PRL, 100 µg), piperacillin/tazobactam (PTZ, 10/100 µg), ticracilin (TC, 75 µg), amikacin (AK, 10 µg), gentamicin (GM, 10 µg), tobramycin (TN, 10 µg), ertapenem (ETP, 10 µg), imipenem (IMI, 10 µg), meropenem (MEN, 10 µg), ciprofloxin (CIP, 5 µg), and cotrimoxazole (TS, 25 µg). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains in each test run.
3.4. Detection of Class 1 and 2 Integrons
DNA extraction was carried out by boiling as mentioned above. PCR amplification of int1 and int2 genes was carried out using two sets of primers listed in Table 1 (16). PCR reaction mixtures (25 µL) contained 0.5 µL of DNA template, 1.5 mM MgCl2, 0.4 mM of each dNTP, 10 pM of primer, and 1 U of Taq DNA polymerase in the buffer provided by the manufacturer (CinnaGen, Iran). The amplification for int1 was performed with the following program: initial denaturation at 95°C for 5 minutes, 30 cycles of 1 minute at 94°C, 1 minute at 54°C, and 1 minute at 72°C, followed by a 10 minutes final elongation step at 72°C. PCR amplification for int2 was carried out at 95°C for 5 minutes, 30 cycles of 30 seconds at 94°C, 30 seconds at 52 °C, and 2 minutes at 72°C, followed by 7 minutes final elongation at 72°C. PCR products were separated on 1% agarose gels and observed as mentioned earlier. Positive controls for class 1 and 2 integron DNA were obtained from the Pasteur Institute in Tehran and were included in the PCR experiments.
3.5. Statistical Analyses
Bivariate analyses were performed to assess correlations between the study variables using non-parametric Spearman’s rank correlation test in SPSS version 22.
4. Results
Biochemical tests and presence of blaOXA-51 gene in all test isolates confirmed their identity as A. baumannii. All isolates (100%) were resistant to aztreonam, amoxicillin-clavulanic acid, ceftazidime, cefotaxime, cefepime, carbenicillin, piperacillin, piperacillin-tazobactam, ticarcillin , amikacin, ertapenem, imipenem, meropenem, ciprofloxacin, and cotrimoxazole. The resistance rates to the remaining antibiotics were: tobramycin (94%), ampicillin-sulbactam (90%), and gentamicin (86%). These results showed that our A. baumannii isolates were extensively drug-resistant (XDR). No correlation could be observed between antibiotic resistance, the source of the specimens, and patient’s gender. Integron carriage was observed in 32 isolates (64%) of which, 11 (34%) harbored class 1 integron, 13 (41%) carried class 2 integron, and eight (25%) had both integrons. No correlation was found between integron carriage and antibiotic resistance phenotype since all isolates were XDR.
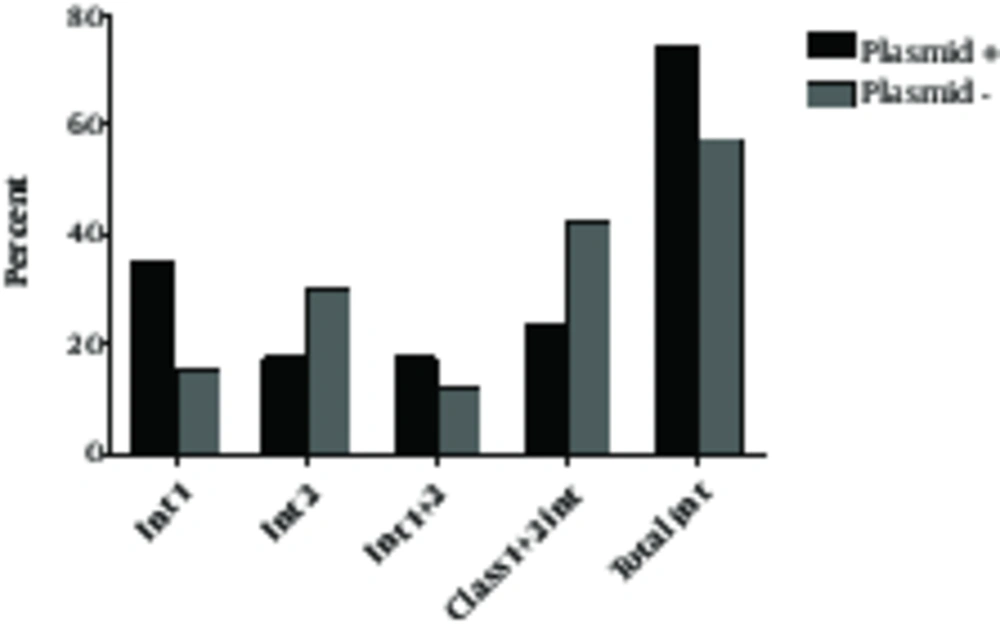
Plasmid carriage occurred in 17 isolates (34%) among which, six (35.4%) carried class 1, three (17.6%) harbored class 2, four (23.5%) had both integron classes, and 4 (23.5%) did not have either of the integron classes (Table 2). Within the plasmid negative isolates, 14 (42.4%) had no integrons, 10 (30.3%) carried integron 2, five (15.2%) harbored integron 1 and interestingly, four (12.1%) carried both integron classes (Table 2, Figure 1). There was a significant association between plasmid carriage and presence of class 1 integron (P = 0.03), but no association was observed between class 2 integron and plasmid carriage (P > 0.05). These results suggest that class 1 integron was largely plasmid-mediated in our isolates. On the other hand, class 2 integron was not associated with plasmid carriage.
| Number of isolates | Number of Isolates (%) Harboring Integron | |||
|---|---|---|---|---|
| Class 1 | Class 2 | Class 1 + 2 | No Integron | |
| Plasmid positive (17) | 6 (35.4) | 3 (17.6) | 4 (23.5) | 4 (23.5) |
| Plasmid negative (33) | 5 (15.2) | 10 (33.3) | 4 (12.1) | 14 (42.4) |
| Total (50) | 11 (22) | 13 (26) | 8 (16) | 18 (36) |
5. Discussion
In this study, all 50 A. baumannii clinical isolates were extensively drug-resistant. Integron carriage was observed in 32 isolates (64%) among which, 34% carried plasmids with molecular sizes of > 20 kbp. The rate of class 2 integron carriage was higher than the rate of class 1 integron and there was a correlation between the presence of class 1 (but not class 2) integron and plasmid carriage. According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the United States Food and Drug Administration (FDA), the criteria for defining XDR in Acinetobacter spp. are non-susceptibility to > 1 agent in all but < 2 categories of the 17 suggested antimicrobial agents (18). We found that all our isolates were XDR. Other Iranian studies have reported XDR rates from 24.4% to 100% (19-22). In addition, a more recent study carried out on 105 A. baumannii isolates showed that all were multidrug resistant and 71.4% were XDR (23). A multi-hospital study performed in Iran over a three-year period from 2011 to 2013 has also shown that 91.3% of A. baumannii isolates were XDR (24).
The worldwide presence of class 1 and 2 integrons in clinical isolates of A. baumannii indicates the prevalence of class 1 integron compared to class 2 integron carriage (25-29). An Iranian study by Goudarzi et al. has also shown the prevalence of class 1 integron (74.1%) over class 2 integron (12.5%) in clinical isolates of A. baumannii where the XDR rate was 62.5% (30). However, in this research, like some other studies from Iran, higher rates for class 2 integron were observed compared to class 1 integron carriage in clinical isolates of A. baumannii (31, 32).
The association of class 1 integrons with resistance to a number of antibiotics has been shown among multidrug-resistant pathogens. Nemec et al. reported that class 1 integron was associated with aminoglycoside resistance among pan-European A. baumannii clones (8). Chen et al. found an association between class 1 integrons and resistance to multiple antibiotics in MDR isolates of A. baumannii (26). Among the reports from Iran, Mirnejad et al. showed a significant correlation between integron carriage and resistance to quinolones, new generation cephalosporins, aztreonam, and amikacin (32). On the other hand, Rastegar-Lari and coworkers did not find any correlation between class 1 integron carriage and antibiotic resistance in burn isolates of MDR A. baumannii (33). We also could not find any correlation between integron carriage and resistance to a specific antibiotic due to the high rates of antibiotic resistance. A recent study by Ramirez et al. showed a class 2 integron in A. baumannii carrying a carbenicillin-resistance gene (blaCARB-4) together with 6 additional resistance determinants comprising four families of antibiotics (34). Few reports have been documented concerning the association between plasmids and integron carriage in A. baumanni. Koeleman et al. found no correlation between plasmid carriage and presence of integrons (35). On the other hand, a study from Poland showed that the class 1 integrase gene was located on a 60 kbp plasmid in an A. calcoaceticus-baumannii complex isolate (28). Our PCR experiments employing both total DNA extracts from A. baumannii isolates and isolated plasmid templates showed a significant association between the presence of a > 20 kbp plasmid and class 1 integron carriage. However, class 2 integron was not associated with plasmids and therefore, it most probably would be located on the bacterial chromosome. Carriage of class 1 integrons by plasmids shows the important role of these mobile genetic elements in the dissemination of drug resistance in A. baumannii.