1. Background
Clostridium difficile are gram-positive spore-forming anaerobic bacteria that are considered as the causative agent for 15% to 25% of antibiotic associated diarrhea (AAD) and also pseudomembranous colitis (PMC) (1, 2).
The majority of C. difficile strains produce two cellular cytotoxins, TcdA, and TcdB; however, a small percentage of pathogenic strains produce truncated nonfunctional toxins. These genes are located within a 19.6-kb genomic pathogenicity locus (PaLoc), where their expression are regulated by tcdC and tcdD. A third toxin, the C. difficile binary toxin (CDT), which functions independently of PaLoc-associated regulatory elements (3, 4), has been identified in approximately 5% of the clinical isolates. Severity of diseases caused by C. difficile mainly depends on types of these virulence genes and their combination in each strain.
Although most cases of C. difficile infection (CDI) occur by endogenous strains after antibiotic therapy, numerous hospital outbreaks were caused through patient-to-patient contact, and contact with the hospital environment and health care workers. Detection of the main sources of infection at hospital wards that are at higher risk of CDI is important for both disease control and its prevention.
Several techniques, such as pulsed field gel electrophoresis (PFGE), restriction fragment length polymorphism (RFLP), arbitrarily primed PCR (AP-PCR), PCR ribotyping, toxinotyping, and enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) have been developed to investigate epidemiology of CDI at the local level (2, 5). However, some techniques may lack sufficient discriminatory power and misclassify the strains. PCR-ribotyping, which is based on size variation of intergenic spacer region (ISR) of ribosomal operon, is the standard typing method in Europe as well as Australia (5-8). Copy number of rRNA (rrn) operons in C. difficile usually range between 1 and 11 (4). Variation within the PaLoc region (containing toxins A and B) and other virulence genes among C. difficile strains has been observed frequently among C. difficile strains and this variation is used for distinguishing different strains from each other (9, 10). This technique is used in two conventional and automated (capillary electrophoresis) manners. Furthermore, diversity in the number of intergenic consensus sequences as palindromes of 127 bp among different strains, which is known as enterobacterial repetitive intergenic consensus (ERIC) sequences, proposed ERIC-PCR as a simple useful tool for molecular epidemiology of CDI (11).
In an attempt to study contamination of medical equipment with C. difficile strains in a gastroenterology unit, the current research compared sensitivity of four methods, PCR-ribotyping, capillary electrophoresis-ribotyping, random amplified polymorphic DNA (RAPD)-PCR assay and ERIC-PCR to determine possible sources of cross-contamination between patients and the environment. Furthermore, consistency of these molecular typing methods was evaluated to determine more simple and suitable typing methods for differentiation of C. difficile strains.
2. Methods
2.1. Bacterial Strains
A total of 23 C. difficile strains from fresh stool samples of patients subjected to colonoscopy (19/105, 18.1%), medical device (3/36, 8.3%), and environmental samples (1/17, 5.8%) of a gastroenterology unit were used for molecular typing. The isolates were obtained and identified as described previously (12). All the samples were collected from a teaching hospital in Tehran, Iran, during December 2010 to August 2011. To show possible correlation, isolates of the medical device and environmental samples were obtained at the same time points. The C. difficile strains were grown anaerobically (Anoxomat, Netherlands) on C. difficile medium (Mast, United Kingdom) supplemented with 7% horse blood and selective components at 37°C. Identity of all isolates was characterized by the polymerase chain reaction, using specific primers (13). This study was approved by the Ethical Committee of Research Institute for Gastroenterology and Liver Diseases (Number RIGLD 613) in Shahid Behehsti University of Medical Sciences, Tehran, Iran.
2.2. DNA Preparation and Polymerase Chain Reaction Assay
Genotyping of C. difficile strains for enterotoxigenic genes tcdA and tcdB, and cytolethal distending toxin (binary toxins, cdtA and cdtB) was done using specific primer pairs, as described by Spigaglia and Mastrantonio (14). Accordingly, crude DNA was extracted from the grown colonies by the InstaGene matrix extraction kit (Bio-Rad, USA) (8). The PCR-amplified products were detected by 1.2% ethidium bromide stained agarose gel electrophoresis.
2.3. PCR-Ribotyping
PCR-ribotyping was conducted in 25-µL reaction volumes composed of 1.5 mM MgCl2, 10 mM Tris-HCl, 50 mM KCl, 2.5 U Taq DNA polymerase, 200 mM of each dNTP, 50 pmol of each primer, and 1 µL of each DNA template. Primers P3 (5’-GCGCCCTTTGTAGCTTGACC-3’) and P5 (5’-CTGGGGTGAAGTCGTAACAAG-3’) were used for the PCR amplification (Ltd., Shiga, Takara Shuzo Co.). The following time-temperature profile was used consistently for each sample: One cycle of five minutes at 94°C for initial denaturation; 35 cycles of one minute at 94°C, one minute at 55°C, and one minute at 72°C; and a final extension cycle of five minutes at 72°C. The amplification products were concentrated to a final volume of 25 µL by heating at 75°C for 20 minutes before electrophoresis, and visualized as described above. The images were analyzed by the Gel ComparII image analysis software (version 3.5, Applied Math) (15). Capillary electrophoresis (CE) ribotyping was performed according to the new consensus ribotyping protocol (12) at the Department of Medical Microbiology, Motol University Hospital, Prague, Czech Republic. The obtained CE-ribotyping profiles were compared with the profiles used in CE-ribotyping validation study (12). The peaks in the chromatogram files (fsa file format) were also uploaded to the WEBRIBO database (16).
2.4. RAPD-PCR Assay
The RAPD-PCR was performed with a single arbitrary primer, 1254 (5’-CCGCAGCCAA-3’). A volume of 25 μL containing 1X PCR buffer, 1.25 µM of each primer, 7 μL of genomic DNA, 200 µM of dNTPs mix, 2.5 mM of MgCl2, and 0.05 U/μL Taq DNA polymerase was used for each reaction. The RAPD-PCR amplification was performed in an automated thermal cycler (AG 22331; Eppendorf, Hamburg, Germany) under the following conditions: Four cycles of five minutes at 94°C, one minute at 36°C, two minutes at 72°C followed by 35 cycles of one minute at 94°C, one minute at 36 °C, and two minutes at 72°C. Similarity of all RAPD banding profiles was analyzed by the GelCompar II Software. Polymorphisms of ≤ 2 and > 2 RAPD bands were considered as definitive criteria for detection of related and different strains, respectively.
2.5. ERIC-PCR Fingerprinting
The primers ERIC1R (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and ERIC2 (5’-AAGTAA GTGACTGGGGTGAGCG-3’) were used for the PCR amplification (17). All PCR amplifications were performed in 25-μL volumes containing 3 μL of template DNA, 1 mM concentrations of deoxynucleoside triphosphates, 2.5 μL of 10X PCR buffer, 1 mM MgCl2, 1 μM concentrations of each forward and reverse primer, 2 U of Taq DNA polymerase at the following conditions: Initial denaturation step at 95°C for seven minutes, followed by 30 cycles of denaturation at 95°C for 30 seconds, 52°C annealing for one minute, and extension at 65°C for eight minutes. After the last cycle, the mixture was incubated at 68°C for 16 minutes (17). The amplification product was analyzed by electrophoresis on a 1.8% agarose gel by a standard protocol after staining with ethidium bromide and visualization on a gel documentation system. Similarity of all ERIC-PCR banding profiles was analyzed by the GelCompar II Software. Relatedness of the strains was characterized in a similar manner that was described for the RAPD-PCR method.
2.6. Statistical Analysis
For analysis of discriminatory power, the current researchers used an index of discrimination for in silico simulation of molecular biology experiments (Discriminatory Power Calculator). Confidence intervals for D were determined by the method described previously by Grundmann et al. (18). Significant differences of the studied assays were estimated using Chi-squared and Fisher’s exact tests.
3. Results
To study possible homology of the C. difficile strains responsible for nosocomial diarrhea, the researchers isolated 19 C. difficile strains from symptomatic patients, (nine males and ten females) and four isolates from medical devices and the environment of a gastroenterology unit (Colonoscope, forceps of the endosonographic imaging system, forceps of the ERCP device and patients’ bed during the study period). Strain RIGLD-141 was used as the control strain for all the conventional microbiological and molecular experiments.
3.1. Toxin Genotyping
Genotyping of the strains for tcdA and tcdB showed one strain as tcdA-/tcdB- (4.34%) and 22 strains as tcdA+/tcdB+ (95.65%). The studied strains also showed cdtA-cdtB- (78.26%; 18/23) and cdtA+cdtB+ (21.73%; 5/23) genotypes for binary toxin encoding genes (Table 1).
| ID code | Source | tcdAB | cdtA | cdtB | CE-Ribotyping | PCR-Ribotyping | RAPD | ERIC PCR |
|---|---|---|---|---|---|---|---|---|
| 32-2 | Colonoscopy device | AB | Neg | Neg | 150/AI-12 | D | VII | J |
| 114-2 | Patient | AB | Neg | Neg | 150/AI-12 | C-Like | XII | H |
| 184-2a | Patient | ABa | Nega | Nega | 001a | B | XIVa | C-like |
| 75-2a | ERCP device | ABa | Nega | Nega | 001a | A | XIVa | B |
| 54-2a | Endoscopy device | ABa | Posa | Posa | 126a | Ba | No pattern | M |
| 65-2a | Patient | ABa | Posa | Posa | 126a | Ba | X | D |
| 20-2 | Patient | AB | Pos | Pos | 126 | E | No pattern | K |
| 141-2 | Patient | AB | Pos | Pos | 126 | F | IX | E-like |
| 85-2 | Patient | AB | Pos | Pos | 126 | G | VI | N-like |
| 105-2 | Patient | AB | Neg | Neg | Unknown | C | VIII | I |
| 90-2 | Patient | AB | Neg | Neg | N/Ab | H | IV | N |
| 83-2 | Patient | AB | Neg | Neg | N/Ab | I | V | A |
| 60-2- | Patient | AB | Neg | Neg | Unknown | J | VIII like | N-like |
| 45-2a | Patient | Neg | Neg | Neg | 039 | Ba | C | Fa |
| 100-2a | Patient | AB | Neg | Neg | 081 | Ba | II | Fa |
| 127-2 | Patient | AB | Neg | Neg | 103 | K | XI | A-like |
| 40-2 | Bed | AB | Neg | Neg | Al- 29 | L | I | K-like |
| 142-2 | Patient | AB | Neg | Neg | 003 | M | No pattern | K |
| 30-2 | Patient | AB | Neg | Neg | Unknown | N | B | M-like |
| 130-2 | Patient | AB | Neg | Neg | N/Ab | B | No pattern | E |
| 134-2 | Patient | AB | Neg | Neg | 029 | B | No pattern | G |
| 110-2 | Patient | AB | Neg | Neg | 017 | O | III | L |
| 89-2 | Patient | AB | Neg | Neg | 014 | B | XIII | C |
| No. of types | 3 | 10 | 15 | 15 | 14 | |||
aNames of the strains that presented the same molecular types by at least three different methods are indicated. Internal nomenclature was used for ERIC-PCR, conventional-PCR, and RAPD-PCR patterns obtained in this study after analysis by GelComparII. The CE-ribotypes were obtained from the WEBRIBO database.
bN/A: not available.
3.2. RAPD Typing
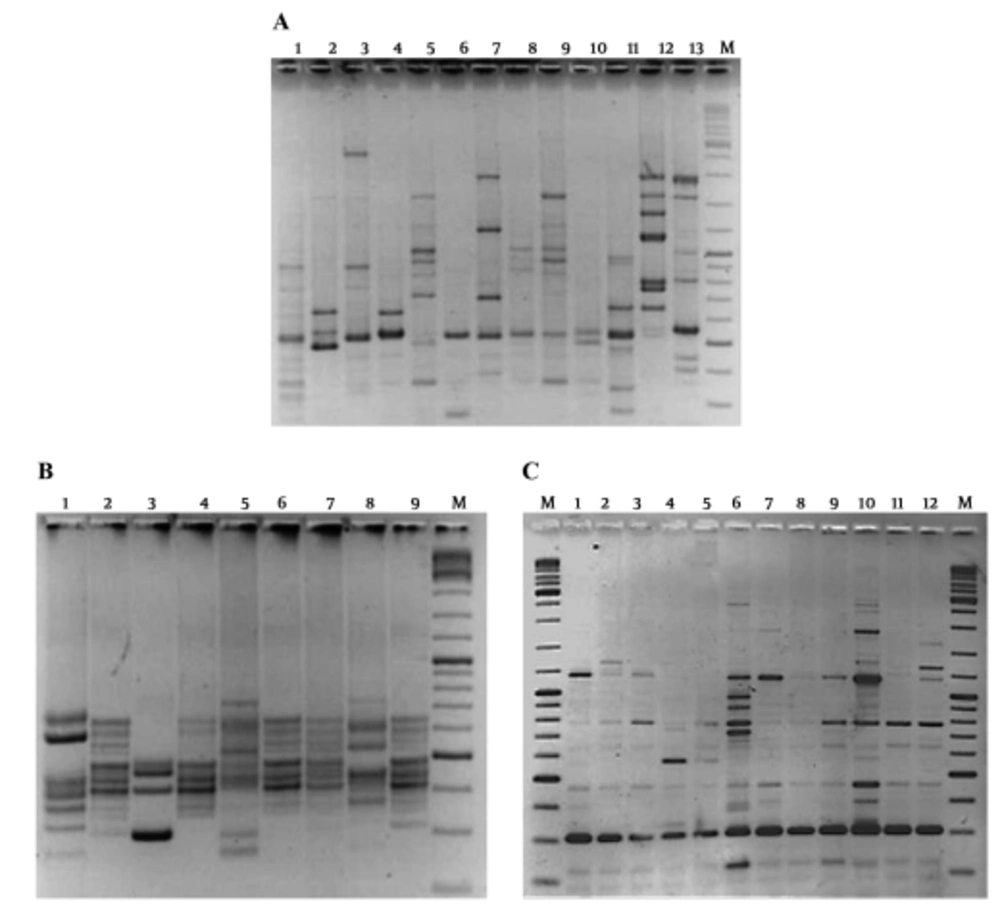
The RAPD-PCR was performed on crude DNA extracts of the C. difficile strains for determination of their genetic relatedness. The PCR results for primer 1283 showed its limitation for discrimination of different C. difficile strains in production of useful banding patterns. In case of primer 1254, five strains did not generate the RAPD patterns; however, 17 different strains were characterized based on their RAPD types (Figure 1A). Cross contamination between the imaging equipment and the patients was shown for the strains presenting RAPD patterns XIV (supplementary file appendix 1). Similarity of RAPD-PCR patterns are shown in Figure 1A. The RAPD typing showed high discriminatory power and stability for analysis of these strains (Table 2).
| Method | D | 95% CI |
|---|---|---|
| Conventional ribotyping | 0.87 | 0.4 - 1.6 |
| CE-ribotyping | 0.89 | 0.8 - 1.2 |
| RAPD | 0.98 | 0.70 - 0.85 |
| ERIC PCR | 0.93 | 0.57 - 1.33 |
Abbreviations: CI, confidence interval; D, discriminatory power.
3.3. PCR-Ribotyping
Results of the conventional PCR ribotyping method showed that all, but one strain, were typeable by this assay. The obtained ribotype patterns exhibited seven to ten bands ranging from 300 to 1100 bp (Figure 1B). A total of 15 different ribotype patterns were found among the 23 studied strains. The predominant type was ribotype B, which belonged mostly to the strains from the patient’s fecal samples. Cross contamination of C. difficile strains between the patients and also medical equipment was seen in strains presenting ribotypes B (Figure 1B and Table 1). All the other strains were characterized as singletons and represent unique ribotypes. Similarity between ribotyping patterns are shown in Figure 1.
(A) RAPD-PCR typing; M: DNA Ladder mix; lanes 4 and 5: Gastroenterology device; lane 2: positive control; lanes 1, 3, and 6 - 13: Patient’s samples; (B) Ribotyping pattern; M: DNA Ladder mix; lanes 7 and 8: Gastroenterology device; lanes 1 - 3 and 9: Patient’s samples; (C) ERIC-PCR results; M: DNA Ladder mix; lanes 7, 8: Gastroenterology device; lanes 1 - 6 and 9 - 12: Patient’s samples.
Comparison of PCR-ribotypes of C. difficile isolates from patients’ fecal samples and gastroenterology imaging device. Homology and diversity of PCR-ribotypes of C. difficile strains were analyzed by GelCompar II software. Correlation of the obtained patterns with toxin gentotypes and RE patterns is shown in the table. Dash line represents homology of five strains (100-2 [patient], 134-2 [patient], 45-2 [patient.], 54-2 [Endoscopy device], and 130-2 [Patient]) based on the obtained ribotype patterns.
3.4. CE-Ribotyping
Results of CE-ribotyping showed ten different ribotypes based on the CE-profiles that were recognized by WEBRIBO. Three CE-profiles were not identified and two DNA samples were not repeatedly amplified by the primers used for this assay.
3.5. ERIC-PCR Fingerprints
The ERIC-PCR fingerprints consisted of patterns comprised of 7 to 12 bands, with the size of the bands varying from approximately 190 to 1500 bp. Two bands (approximately 300 and 480 bp) were conserved among all isolates (Figure 1C). While the strains showed two identical (F and K) and six related (C, E, N, A, K, and M) ERIC types, the strains provided 14 different patterns in total (supplementary file appendix 2).
3.6. Correlation of Ribotyping, Toxin Genotyping, and RAPD-PCR
Comparison of the results of all the molecular typing methods showed some diversity. Congruency of the results among the assays was observed for six strains. Two strains that showed CE-ribotype 001 (C. difficile isolates from ERCP and a patient) also presented similar RAPD and virulence types. Furthermore, C. difficile isolates from endoscope and a patient showed the same genotype, CE-ribotype and conventional ribotype (126) (Table 1). While consistency of results of ERIC-PCR and PCR ribotyping was detected for two patients isolates (45-2 and 100-2), further analysis showed their difference in CE-ribotype and virulence genotype patterns. Results of Simpson’s index diversity showed RAPD-PCR as the method with highest discrimination ability and conventional PCR-ribotyping method as the one with lowest discrimination ability.
4. Discussion
During the past 20 years, spore forming C. difficile were characterized as highly-resistant microorganisms to commonly used antibiotics and disinfectants (19). Outbreaks of CDAD (C. difficile associated disease) have emerged due to the prescription of broad spectrum antibiotics in hospitalized patients, which mainly rely on the alteration of the intestinal microbiota and overgrowth of the colonized spores or resistance strains in this tissue (20, 21). Infection with these bacteria could be caused by both endogenous and exogenous routes (19). Application of inappropriate disinfected medical devices, admission of patients to hospitals with poor health services, and usage of contaminated medical foods are the main risk factors for acquisition of C. difficile in these patients (22). Results of this study showed contamination of the gastroenterology imaging devices. There are a few reports about the recovery of C. difficile from medical equipment and its transmission to patients in hospital settings (23-25). Molecular relatedness of C. difficile strains from medical equipment and patients’ clinical samples was only established in one study by Dumford et al. (24). However, none of these strains were isolated from gastrointestinal imaging devices. According to the author’s knowledge, this is the first report that presented the occurrence of cross contamination of C. difficile between a gastrointestinal imaging device and patients, who were subjected to examination by this equipment. The noted contamination could be explained by non-standard washing and sterilization process of the equipment at the studied hospital (glutaraldehyde 2% and 10 minutes of sterilization), which occurred because of the high number of admitted patients for the examination.
Emergence of hyper virulent strains of C. difficile, BI/NAP1/027, in North America and Europe, has increased attention of physicians for elimination of hospital sources of infection in the recent years (26). Because of the worldwide emergence of virulent strains of C. difficile, epidemiology of this organism and investigation of its role in CDI seems to be important (27, 28). There are a variety of different phenotyping and molecular typing methods that were proposed for epidemiology of CDI (28). Both of the methods have some limitations, however, molecular typing methods provide greater levels of typeability (29). Furthermore, RAPD-PCR and PCR ribotyping, are among commonly applied methods for molecular typing of C. difficile strains in clinical and non-clinical samples (30-34). It was expected for these typing methods to be able to cluster the same isolates in similar genetic profiles (28). However, discriminatory power of these methods for different strains was not clear. Tenover et al. by analyzing a greater numbers of isolates, showed that PCR-ribotyping provides higher levels of strain discrimination than PFGE. Weakness of PCR-ribotyping in discrimination of outbreak strains was described by Killgore et al. However, these authors presented a superior discrimination ability for PCR-ribotyping compared with toxinotyping methods (28). Brazier showed PCR ribotyping, among the other molecular typing methods, as a more discriminative and reproducible method (29). On the basis of the results, it appeared that all the studied techniques were able to type most of the isolates (14 types by the ribotyping method). However, the RAPD-PCR technique showed higher discriminatory power than the others (17 types). The current results are comparable with those reported by Green et al. (32). They showed that PCR ribotyping in conjunction with RAPD-PCR assay categorized different types within defined PCR ribotypes. They showed different RAPD types of C. difficile within the strains with the same PCR ribotype and concluded that combination of PCR ribotyping and RAPD technique could provide a greater discriminatory power than either of the methods when used alone. Van Dijck et al. by studying 56 toxigenic isolates, indicated that combination of the RAPD-PCR technique and PFGE could determine genetic diversity within toxigenic C. difficile isolates (35). Barbut et al. reported on the RAPD assay as a valuable tool for epidemiological studies of C. difficile (36). An excellent correlation was obtained between the results of RAPD and PFGE, and ribotyping in a study by Chachaty et al. for clustering of C. difficile isolates. Their study showed that PFGE and RAPD-PCR had similar discriminatory power (26 different types by PFGE, 25 by RAPD, and 18 types by ribotyping). In case of ERIC-PCR, the current research found that this method had the lowest discriminatory power in comparison with the other methods. In accordance with the current results, Rahmati et al. showed that ERIC-PCR could not discriminate different strains of C. difficile (37).
Molecular relatedness of C. difficile strains from hospital wards and patient samples was only established in a comprehensive study by Dumford et al. Results of this study showed that CDI incidence correlated with the prevalence of environmental C. difficile in hospital wards. In their study, RAPD and RS PCR (RNA template-specific polymerase chain reaction) typing showed similar discriminatory power (38). The presence of C. difficile strains with similar molecular types in the studied patients was comparable with those seen in Europe, particularly the UK (39), Hungary (40), and Poland (41). They showed ribotype 001 as a responsible strain for 55% C. difficile infection in UK hospitals and ribotype 078 for 39% of all the isolates in Hungary. On the other hand, the association of all the environmental isolates and 35% of the neonatal isolates was established in Poland (41). Dominance of some ribotypes and their involvement in nosocomial diarrhea was reported by some studies. In a study by Rotimi et al., two ribotypes were responsible for over one-third of the cases of CDAD in Kuwaiti hospitals (42). In a European survey, among thirty-eight hospitals in fourteen different countries, sixty-six different PCR ribotypes were characterized among 322 toxigenic strains of C. difficile, which was higher than those obtained in the current study (10 ribotypes among 23 strains) (40, 43, 44). In the current experiment, C. difficile CE-ribotype 126 and conventional ribotype B were the most common ribotypes among the studied patients.
4.1. Conclusion
In conclusion, cross contamination of C. difficile between patients and a medical imaging device was established by different molecular typing methods in the current study. Although PCR-ribotyping, CE-ribotyping, RAPD-PCR, and ERIC PCR methods were able to show diversity of C. difficile strains, there was some diversity among these techniques for typing purposes. None of the studied methods was able to detect the identical strains solely. While RAPD-PCR and ERIC-PCR showed the highest discriminative power for differentiation of the C. difficile strains in the current study, their weakness was shown due to inconsistency in comparison to results of the other typing methods. These results proposed usage of a combination of two or more typing methods for better assessment of diversity among them.
![Comparison of PCR-ribotypes of <i>C. difficile</i> isolates from patients’ fecal samples and gastroenterology imaging device. Homology and diversity of PCR-ribotypes of <i>C. difficile</i> strains were analyzed by GelCompar II software. Correlation of the obtained patterns with toxin gentotypes and RE patterns is shown in the table. Dash line represents homology of five strains (100-2 [patient], 134-2 [patient], 45-2 [patient.], 54-2 [Endoscopy device], and 130-2 [Patient]) based on the obtained ribotype patterns. Comparison of PCR-ribotypes of <i>C. difficile</i> isolates from patients’ fecal samples and gastroenterology imaging device. Homology and diversity of PCR-ribotypes of <i>C. difficile</i> strains were analyzed by GelCompar II software. Correlation of the obtained patterns with toxin gentotypes and RE patterns is shown in the table. Dash line represents homology of five strains (100-2 [patient], 134-2 [patient], 45-2 [patient.], 54-2 [Endoscopy device], and 130-2 [Patient]) based on the obtained ribotype patterns.](https://services.brieflands.com/cdn/serve/3170c/cb3c940d919e502f162c486dca6edf5de3fd5f85/archcid-13-5-61030-i002-preview.webp)