1. Background
Non-Tuberculosis Mycobacteria (NTM) are environmental organisms that can cause opportunistic infections in humans. While many NTM species are considered as free-living saprophytes, an increasing number have been reported as opportunistic pathogens, which are capable of causing serious diseases in humans (1, 2). In the last decade, the prevalence of NTM has been increasing worldwide and NTM accounts for an increasing proportion of mycobacterial diseases (2-4). Furthermore, NTM infections are similar to tuberculosis (TB) in their clinical and microbiological presentation and may cause a clinical dilemma with regards to therapy for infected patients (5). In Iran and most other developing countries, in which TB is an endemic disease, diagnosis of TB is difficult and clinicians mostly rely on conventional tests (i.e. tuberculin skin testing, chest radiography, and microscopy examination), a practice that may not distinguish NTM from TB (4, 6). A recent report from Iran found that 30% of patients with NTM infections were wrongly diagnosed as MDR-TB (6).
2. Objectives
Given the above facts, this study aimed at reporting the prevalence of NTM among TB suspects, referred to a teaching hospital in Tehran, Iran.
3. Methods
3.1. Samples and Setting
A total of 230 clinical isolates from TB suspected cases that were referred to Baqiyatallah hospital from March 2016 to July 2017 were included in this study. This hospital has well-equipped biosafety level III laboratory facilities and standard safety precautions were followed for specimen processing. The ethics committee of Baqiyatallah hospital approved the study and all the patients signed an informed consent.
3.2. Isolation and Identification of Mycobacteria
Obtained specimens were processed using Petroff’s method (7). The obtained sediments were stained with Ziehl-Neelsen and were cultured in Lowenstein-Jensen medium (7). Culture media was examined twice per week for growth rate, colony morphology, and pigmentation, according to standard procedures (8).
3.3. Identification of M. tuberculosis
GeneXpert assay was performed for assignment of isolates to M. tuberculosis, as described previously (9, 10). Briefly, sample reagent was added at a 2:1 ratio to clinical specimens and then the mixture was added to the Xpert MTB/RIF cartridge to perform the assay in the Genexpert instrument. The automatically generated results were read after 90 minutes.
3.4. Identification of NTM Species
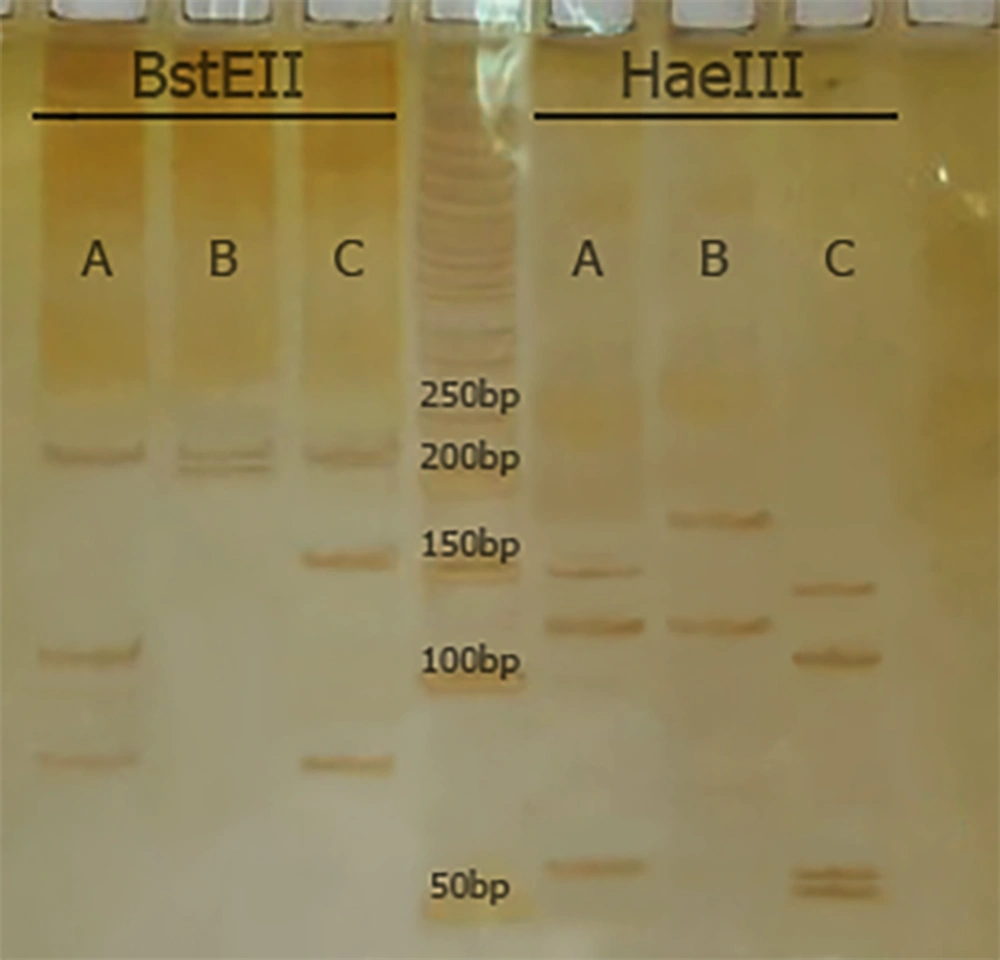
Polymerase Chain Reaction restriction analysis (PRA) and hsp65 gene sequencing were used to speciate NTM. For PRA, a set of primers (Tb11 and Tb12) was used to amplify a 440-bp fragment (Tb11: 5’-ACCAACGATGGTGTGTCCAT-3’, and Tb12: 5’-CTTGTCGAACCGCATACCCT-3’). The Hae III and Bst II restriction enzymes were used to digest the amplified products and electrophoresis was performed on 8% polyacrylamide gel (11). The algorithm proposed by Roth et al. was used for species identification (12).
For sequencing, the amplified PCR products of hsp65 gene for each isolate were sequenced using specific primers, as described previously (11).
4. Results
4.1. NTM Isolation
Of 230 culture positive TB isolates, 12 isolates (5.2%) were NTM. The remaining, 218 isolates were confirmed as M. tuberculosis using the GeneXpert method.
4.2. PRA and hsp65 Sequencing
According to PRA and hsp65 sequencing, M. simiae (6, 50.0%), M. fortuitum (4, 33.3%), M. intracellulare (1, 8.3%), and M. kansasii (1, 8.3%) were the most commonly isolated NTM, respectively (Figure 1 and Table 1). Clinical isolates were confidently identified by hsp65 gene sequencing.
| Numbers of Isolates | Phenotypic Tests | Patterns by hsp65-PRA | Identification by PRA | hsp65 Sequencing | |
|---|---|---|---|---|---|
| Bst E II | Hae III | ||||
| 6 | Mycobacterium sp. | 235/210 | 185/130 | M. simiae | M. simiae |
| 4 | Mycobacterium sp. | 235/120/85 | 145/120/60/55 | M. fortuitum | M. fortuitum |
| 1 | Mycobacterium sp. | 235/120/100 | 145/130/60 | M. intracellulare | M. intracellulare |
| 1 | Mycobacterium sp. | 235/210 | 130/105/80 | M. kansasii | M. kansasii |
5. Discussion
In Iran, diseases caused by NTM appear to be increasing and represent an important public-health threat. Although knowledge about NTM diseases seems to be improved, yet reliable diagnosis of NTM still remains problematic.
According to the current study, 5.2% of isolates, which were obtained from TB-suspected patients were positive for NTM. Several reports suggest that there is an increasing number of NTM worldwide (13-19). The current researchers have recently shown a relatively high prevalence of NTM infections (10.2%) among culture-positive cases of TB, which emphasizes on the role of NTM in public health (4).
Mistaking patients with NTM for TB is a serious error (6). So far, all NTM are acid-fast; clinical manifestations of many NTM are often indistinguishable from TB and disease caused by NTM, usually does not respond to anti-TB drugs (6, 20). Thus, accurate diagnosis of mycobacterial species is very important and can facilitate the prevention and control of NTM infections.
Mycobacterium simiae was the most common NTM species from investigated isolates in the current study. This was similar to previous studies, which were conducted in Iran (21-23). Mycobacterium simiae is an endemic NTM in Iran and its clinical and microbiological findings are often indistinguishable from TB (21, 22). Thus, the relatively high prevalence of M. simiae may cause serious problems for TB control strategies in Iran.
Mycobacterium fortuitum and M. kansasii were also among the most common NTM species, which were isolated from suspected TB patients. Similarly, Hashemi-Shahraki et al. found that M. fortuitum and M. kansasii were the most common cause of NTM diseases in Ahvaz (a city located in the south of Iran) (19). They evaluated 92 isolates from patients with confirmed NTM diseases and concluded that PRA and sequencing are reliable methods for elucidating taxonomic data and species identification of Mycobacterium isolates (19).
The first limitation of the current study was the relatively small clinical sample size, thus, this research have underestimated the true prevalence of NTM. Secondly, although the current patients were likely to have pulmonary NTM disease, yet the researchers were unable to completely rule out the possibility of NTM contamination of the positive cultures. In conclusion, the relatively high isolation of NTM strongly highlights the need to make strategies for surveillance, monitoring and management of NTM cases.