1. Background
Enterococci constitute an important and diverse group of bacteria and have a complex association with human beings. Some of these species are used in food industry, while some others can cause various diseases in humans and animals (1). Enterococci are Gram-positive bacteria constituting normal flora in birds, humans, and animals intestines, and they are found in the mouth cavity, intestine, and vagina of humans. They are the most abundant Gram-positive cocci in human faeces (2). These microorganisms are the most common causes of hospital infections, particularly in ICU, the second agent in causing urine infection in hospitals, and the third agent in hospital bacteremia, endocarditis, and other infections among the ICU patients (1, 3). E. faecalis and E. faecium are 2 common species isolated from nosocomial infections, and among the different species of enterococci that are currently recognized, E. faecalis is responsible for 85% to 95% of enterococcal infections, while 5% to 10% of infections are caused by E. faecium (4). Enterococci are considered as a problematic bacteria in antibiotic treatments, as these bacteria resist against most antibiotics innately and can also acquire resistance through plasmids or transposons (5). Hospital infections by enterococci is the main issue in hospitals, which is increasing in most countries including Iran (6). Due to the inappropriate use of antibiotics in nosocomial infections, the resistance rate is increasing (4). Aminoglycosides are frequently used in combination with cell wall active antibiotics for severe enterococcal infections. Unfortunately, enterococci with high-level resistance to aminoglycosides (HLAR) (MIC > 500 µg/mL) leads to the loss of synergy effect between a cell wall active agent and an aminoglycoside, typically gentamicin (7). Enterococcus species can acquire high-level resistance to a variety of antibiotics by horizontal transfer of mobile genetic determinants in addition to the intrinsic resistance to several groups of antimicrobials (3). Resistance to the aminoglycosides usually occurs by enzymatic modification of the antibiotics by aminoglycoside-modifying enzymes (AME); aac (6’)Ie-aph (2”)Ia is the most common gene causing HLGR in enterococci (8). The first case of high-level gentamicin-resistant (HLGR) E. faecalis isolate was reported by Thal et al. (1979) in France (9). The resultant bifunctional aac (6’)-aph (2”) aminoglycoside modifying enzyme confers resistance to all clinically useful aminoglycosides except streptomycin. The genes responsible for high-level aminoglycoside resistance have been identified on plasmids in most cases (1). Murray and Hodel found that aac (6’) Ie-aph (2”) Ia gene is located on transposon, Tn5281, whose presence on the transposon is a reason for rapid dispersion and high outbreak of HLGR in enterococci and increases the probability of horizontal transfer of the gene. Moreover, this gene is located on transposon Tn4001, which has been assigned to staphylococci, with similar transposon Tn4001 (10). An increase in the prevalence of HLGR has been observed in European, Asian, and South American countries (11).
2. Objectives
Due to the importance of enterococci in hospital infections and their role in human health, the antibiotic resistance pattern in the enterococci isolated from a pediatric hospital and presence of aac (6’) Ie-aph (2”) Ia gene in HLGR enterococcal isolates were investigated in this research to obtain information on antibiotic resistance.
3. Methods
3.1. Sample Collection and Phenotypic Identification
This cross-sectional, descriptive, and prospective study was conducted on 100 Enterococcus species (50 clinical isolates and 50 faeces isolates) collected during September 2014 and June 2015 from a pediatric hospital in Tabriz, northwest of Iran. To isolate enterococci, the M-Enterococcus agar (Quelab, Canada) was used; after culturing and 48- hour incubation at 37°C (Memmert, Germany), the enterococci typical colonies were cultured on the blood agar (Biolife, Italy) plates and incubated for 24 hours at 37°C. After preparing a pure culture of the isolates, they were investigated to determine their genus and species. The genus was identified by Gram’s staining, catalase test, growth in presence of 6.5% NaCl, and hydrolysis of bile esculin. To identify the enterococci species, the biochemical reactions of arabinose and sorbitol fermentation and motility tests were employed (12).
3.2. Determination of Antibiotic Susceptibility
Antibiotic susceptibility testing of bacterial isolates was performed using Kirby-Bauer disk diffusion method and Mueller-Hinton agar (Merck, Germany). Bacterial suspension equal to a 0.5 McFarland turbidity standard and the diameter of the inhibition zone around the disk based on mm were recorded after incubation for 24 hours at 35°C, according to CLSI (Clinical and Laboratory Standards Institute) (13). Antibiotic disks were high dose of gentamicin (120 μg), ampicillin (10 μg), vancomycin (30 μg), erythromycin (15 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), and tetracycline (30 μg) (Biomaxima - Poland). Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were used as a control strain for the susceptibility testing.
3.3. DNA Extraction
DNA extraction in HLGR isolates was performed by tissue buffer. First, NaOH 0.4 g and SDS 0.5 g were dissolved in the 200 μL deionized water. Then, 20 μL tissue buffer was added to micro tubes and some colonies were removed from fresh culture (18 - 24 h) and dissolved in the micro tubes containing tissue buffer and placed at 95°C for 10 minutes; they were then centrifuged by 13 000 rpm (vision- South Korea) for 1 minute, and the supernatant was removed by sampler and placed in a new micro tube. Finally, 180 μL of deionized water was added to each micro tube and stored at -20°C for PCR reaction (14).
3.4. Polymerase Chain Reaction
PCR was performed to amplify the aac (6’) Ie-aph (2”) Ia gene. Primers 5’CAGGAATTTATCGAAAATGGTAGAAAAG3’ and 5’CACAATCGACTAAAGAGTACCAATC3’ were used as forward and reverse primers to obtain PCR product (17). PCR amplification was performed in a total volume of 25 μL containing 6μL of deionized water, 12 μL of PCR master mix (Yekta Tajhiz Azma, Iran), 2 μL from each primers (Sinaclon, Iran), and 3 μL DNA. Thermal cycling (Eppendorf, Germany) was performed by initial denaturation at 94°C for 3 minutes followed by 35 cycles of 40 seconds denaturation at 94°C, 40 seconds annealing at 57°C, and 40 seconds extension at 72°C with a final extension at 72°C for 2 minutes. PCR products were analyzed by electrophoresis (UVP, Bio Doc, United states) in 1% agarose gel and visualized by gel documentation system after staining with ethidium bromide (15). E. faecalis strain HH22, containing the HLGR-conferring transposon Tn5281 and E. faecalis ATCC 29212 were used as positive and negative controls, respectively (12).
3.5. Data Analysis
The data were analyzed using the chi-square test with SPSS 17 (SPSS Inc., Chicago, Illinois, USA). Significance level was set at P < 0.05. Logistic regression was used to calculate odds ratios (ORs) with 95% confidence intervals (CIs).
4. Results
The isolated enterococci are presented in Table 1 according to place of isolation. Of 50 clinical isolates, 27 (54%) and 23 (46%) were E. faecalis and E. faecium, respectively. Of 50 faeces isolates, 2 (4%) and 48 (96%) were E. faecalis and E. faecium, respectively. Among 100 enterococcal isolates, 31 (62%) were HLGR, 70.96% of which belonged to clinical isolates, and 29.03% to faeces isolates (P < 0.001).
| Specimen | Number | Percent |
|---|---|---|
| Urine | 41 | 82 |
| Wound discharge | 1 | 2 |
| Blood | 5 | 10 |
| Ascites fluid | 1 | 2 |
| Skin tissue biopsy | 1 | 2 |
| Ear discharge | 1 | 2 |
| Total | 50 | 100 |
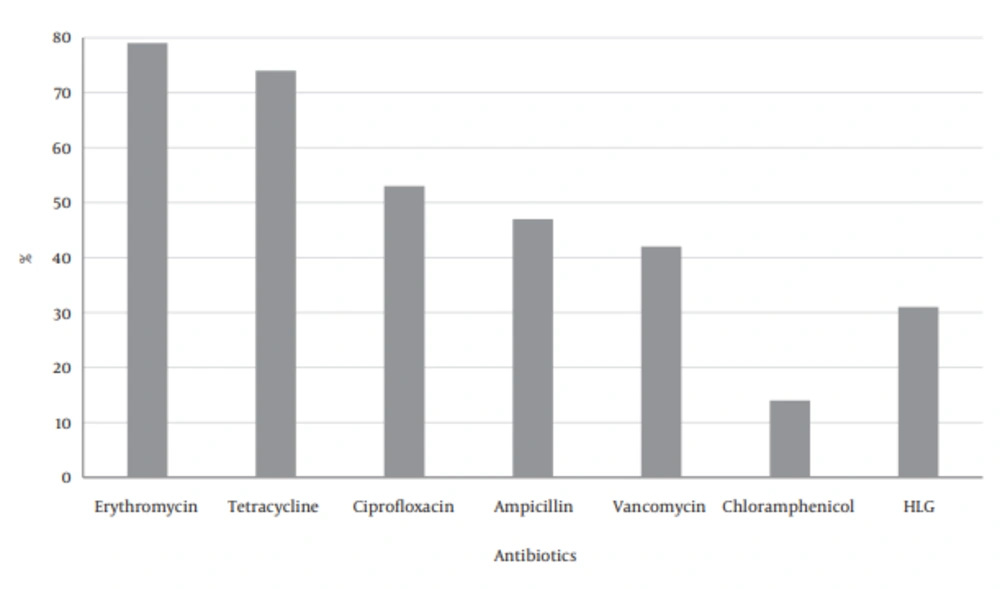
The most resistance against the antibiotics was found in erythromycin and tetracycline, and then ciprofloxacin and ampicillin, followed by less resistance to chloramphenicol (Figure 1).
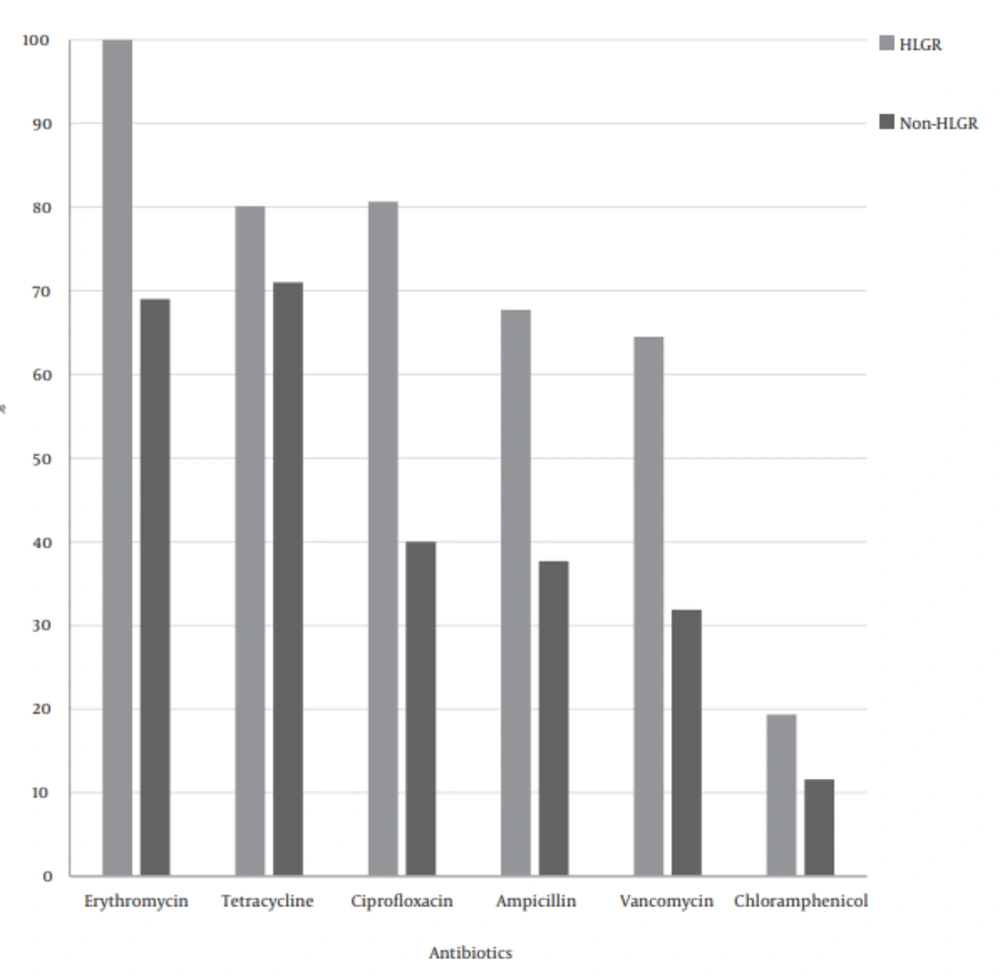
Of all isolates, 48% were resistant to more than 3 different antibiotics, as 23 isolates (46%) of enterococcus isolated from faecal specimens and 25 isolates (50%) of enterococci isolated from clinical specimens were MDR. Multidrug resistance (MDR) rates in HLGR isolates were higher than non- HLGR isolates (Figure 2).
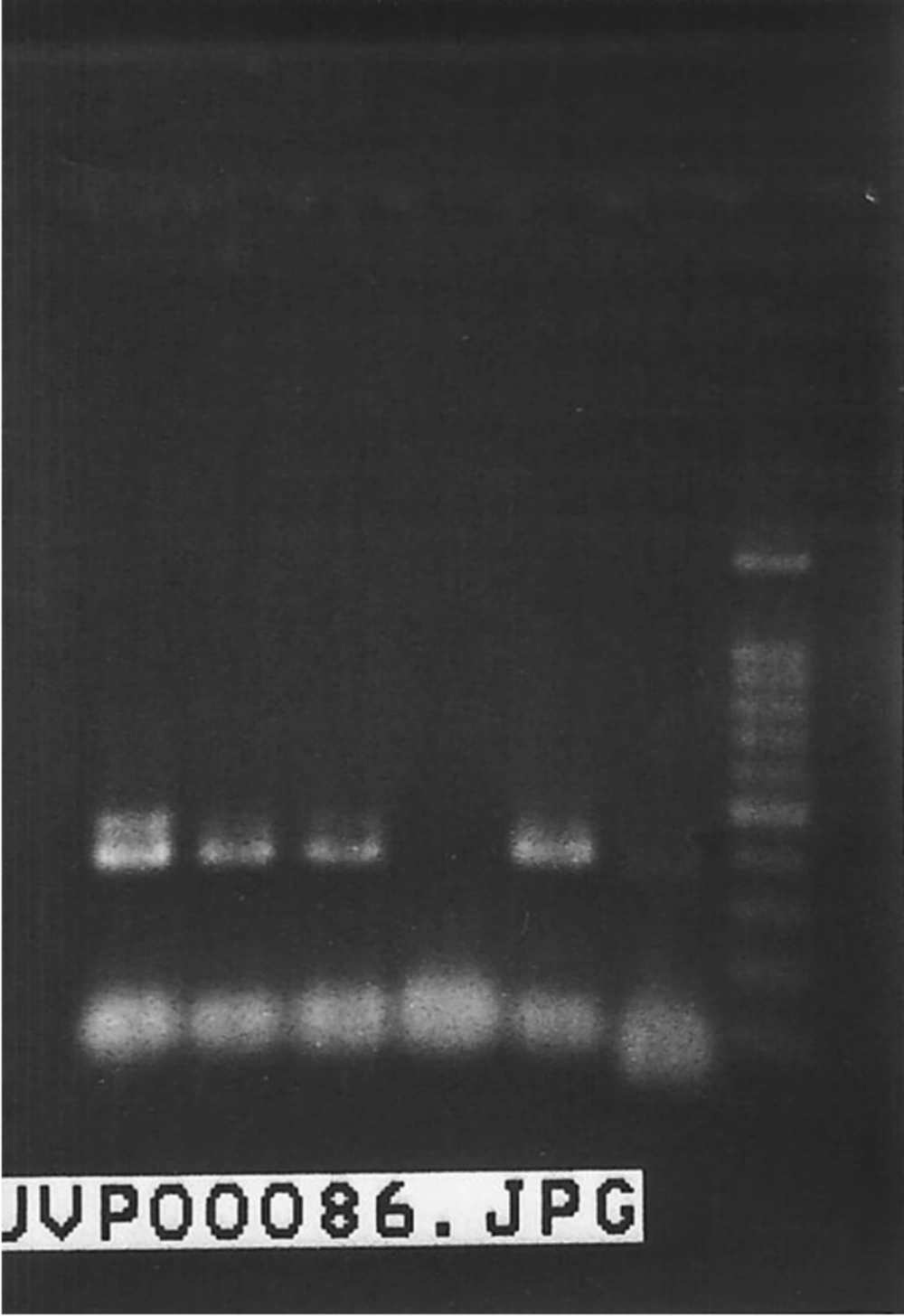
Among 31 isolates that were identified as HLGR, the aac (6’)-Ie-aph (2”)-Ia gene was detected in 21 (67.74%) isolates (P < 0.05). Figure 3 shows a 369 base pair PCR amplicon associated with high level gentamicin resistance.
5. Discussion
In recent years, enterococcus has been known as an important pathogen in Iran like other parts of the world. The increasing number of enterococcal species resistant to antibiotics are currently a challenge with regard to treatment (16). In addition to financial losses caused by treatment of these infections, in studies on enterococci, the mortality rate attributable to enterococcal bacteremia was 31% (17).
Overall, 82% of these Enterococcus strains were isolated from urine samples; therefore, the role of enterococci in urinary tract infections was further illustrated (16).
Among enterococci isolated from faeces, the E. faecium was dominant, and similar results were obtained in Canada and Ethiopia (18, 19). Also, in a research in South Korea (2010) and in India (2011), the most common species was E. faecium with frequency of 70% and 50%, respectively (20, 21).
In our clinical specimens, as expected, E. faecalis isolates showed the highest incidence. In a research by Dadfarma and Behnood, E. faecalis was also the most common species in clinical specimens (8, 22). Species distribution varies in various geographical regions due to different climate and bacterial conditions. In India and Japan, E. faecium had a high percentage of clinical enterococci, but in Iran, US, England, and other European countries, E. faecalis is a common species isolated from clinical specimens (23). According to different studies, diversity of strains can be assigned to the studied samples and time of sampling.
Different results have been obtained regarding the high level of resistance to gentamicin. In this study, 58% of HLGR isolates belonged to E. faecium. Some past studies have also indicated that E. faecium is the main species. In Italy, from the total HLGR isolates, 52% were E. faecium and 44% were reported as E. faecalis (24). In South Korea, 70% of HLGR isolates belonged to E. faecium (20). Also, in a study by Behnood et al. (2013) in Tabriz, it was found that 57.1% of HLGR isolates were E. faecium and 28.84% were E. faecalis (22). In this study, the most common species of HLGR isolates was E. faecium, which depicts the importance of this enterococcus in spreading a high- level resistance to gentamicin by colonization in the hospitalized patients’ intestines and also through hospital and urban sewage system. High level resistance to gentamicin has caused an increase in E. faecium in treatment centers. Although E. faecalis is a pathogen with high pathogenesis, E. faecium due to its high potential in resistance against antibiotics, shows a high percentage of resistance to various antibiotics (24).
In the past 4 decades in Iran, aminoglycosides, particularly gentamicin, have been used broadly in the treatment of infections, which can be one of the causes of high prevalence of HLGR isolates in Iran’s hospitals (25). In our study, high-level resistance to gentamicin was observed in 31 % of isolated enterococci. This frequency is lower than the rates reported by Dadfarma (57.4%, 2013) and Feizabadi (65%, 2008) (8, 26). In this study, the frequency of HLGR isolates was higher in clinical specimens, which may be due to high consumption of antibiotics in clinics or due to collection of samples from hospitalized patients. HLGR was reported in 46.15% of isolates in Italy (24), 45.5% in Brazil (27), 37.64% in Chicago (28), 63% in South Korea (20), and 62% in Delhi (21). These results demonstrate variation of HLGR prevalence in different geographic regions. Resistance to aminoglycosides usually occurs by enzymatic modification of drugs through aminoglycoside modifying enzymes, which are carried on by mobile genetic elements such as transposons. The HLGR in enterococci is usually coded by the aac (6’) Ie-aph (2”) Ia gene (29), and studies on different genes causing resistance to gentamicin in HLGR isolates in Iran and the world have reported high presence of aac (6’) Ie-aph(2”) Ia gene (30). The results of this study revealed that 67.74% of HLGR isolates carried aac (6’)-Ie-aph (2”)-Ia gene. High percentage of this gene in HLGR isolates shows its specific role and high dispersion in Iran. The prevalence rates of resistance to other antibiotics and multi-drug resistance (MDR) was higher among the HLGR isolates compared to the non-HLGR isolates; 90.6% of HLGR isolates and 48% of non-HLGR isolates were MDR. Also, in Dadfarma study, multi-drug resistance was high in HLGR isolates relative to non-HLGR isolates (8).
To the best of our knowledge, this was the first report showing the high-level gentamicin resistance among enterococcal isolates from Tabriz pediatric hospital; however, we could investigate more enterococci from other parts of Iran to find more precise and comprehensive results about their HLGR pattern.
4.1. Conclusion
Our results revealed a high prevalence of MDR and HLGR, which underlines the necessity of control and prevention measures resistance in enterococci through epidemiological investigation of resistance and application of molecular typing methods. Using antibiotic sensitivity disks, particularly aminoglycoside, is essential in antibiotic sensitivity tests. Their antibiotic resistance and molecular typing could help us understand their medically important organisms; moreover, the use of aminoglycoside antibiotic disks in sensitivity testing could help us select an appropriate treatment.