1. Background
Helicobacter pylori (H. pylori), a Gram-negative bacterial pathogen, is recognized as an important risk factor for chronic active gastritis, peptic ulcer, gastric cancer, and gastric mucosa associated lymphoid tissue lymphoma (1-4). Approximately half of the world’s population is infected with H. pylori; however, most of the infected people have no symptoms (3, 5). Although initial colonization of this bacterium occurs in early years of life, occurrence of disease depends on its genetic entity and virulence property in association with host and environmental risk factors (6). Diversity of H. pylori strains was demonstrated in several studies and in different populations (7-10). Association of this diversity with the severity of gastric disorders was shown for some structural and functional proteins of H. pylori (5, 8, 11, 12).
Cag-pathogenicity Island (Cag-PAI) is one of the complex genetic structures in bacteria that encodes approximately 27 to 31 genes (13-15). While involvement of this structure in inflammation of gastric tissue and its role for delivery of CagA oncoprotein into gastric epithelial cells were demonstrated, there is very few data regarding its association with the clinical outcome of the infection (5, 6). At least 17 out of 27 cagPAI genes encode main structural proteins that are important for providing a putative type IV secretion system (T4SS) to represent a needle-like pilus device including CagL, CagY, CagE, CagW, and Cagα. Some of these proteins, especially exterior proteins, including CagT, CagY, and CagX interact with host cell receptors and their presence are essential for delivery of effector protein CagA and peptidoglycan into the cells (16-22).
Diversity of cag-PAIs among different H. pylori strains, encouraged researchers to use cag-PAI typing as a molecular typing method to study its association with their pathogenic capacity in infected patients (23-25). Diversity of this PAI could be screened for the presence or absence of noted genes or defined deletions and insertions. Since there are increasing reports regarding sequence diversity of dedicated genes of cag-PAIs, data regarding the integrity of this structure using cag-PAI typing could not signify its proper function. cagA is considered as a more variable gene of cag-PAI and contains a variable number of repeat sequences located in the 3’ of this gene (known as EPIYA motif) that may affect its biological activity. Recombination of these repeat units during replication may convert it to a less virulent oncoprotein or a pseudogene (5, 26).
Study of clinical isolates of H. pylori, at both molecular and functional levels, could provide more valuable data that somewhat explain the diversity in the clinical outcomes of infected patients. However, such correlation was fairly investigated. This study was aimed to analyze the diversity of H. pylori strains in capacity of CagA expression and its association with EPIYA types, intactness of cag-PAI, and variation in promotion of different gastric disorders in human host tissue.
2. Methods
2.1. Bacterial Strains and Culture Conditions
CagA positive H. pylori strains were randomly selected from the microbial collection of research institute for gastroenterology and liver diseases in Tehran, Iran. All the strains were obtained from patients with gastric disorders. The identity of the strains was confirmed by the culture and polymerase chain reaction (PCR) using species specific primers for gene glmM (27). Briefly, the isolates were recovered from the stocks on Brucella agar medium supplemented with fetal calf serum (10%) (v/v), horse blood (7%), selective supplement (vancomycin 2.0 mg, polymyxin B 0.05 mg and trimethoprim 1.0 mg, Merck, Germany), and amphotericin B (3 mg/L). The plates were incubated at 37°C for 3 to 5 days in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2). The grown colonies were used for biochemical characterization, based on their morphology as well as positive reactions for oxidase, catalase, and urease tests. Genomic DNA of the selected strains was extracted from the harvested colonies, using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. H. pylori strain RIGLD-OC661 was used as a reference strain for all the experiments.
2.2. Screening of cagPAI Gene Loci by PCR
To analyze the integrity of cagPAI among these strains, the presence of cagA was initially determined using specific primers covering the entire 3’ region of cagA gene, cag3F: 5’-TCCGTTAAAGATGTGATCATCAATC-3’and cag3R: 5’-AGATTTTTGGAAACCACCTTTTG-3’ (28). Amplification of cagA was performed as follows: an initial denaturation at 94°C for 4 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 64°C for 30 seconds, and extension at 72°C for 90 seconds, with a final elongation step at 72°C for 10 minutes. CagA EPIYA types was determined based on the size variation of the PCR products in comparison to reference strains for common EPIYA types (Accession numbers JX428788, JX428766, JX428786, JX428784).
All the cagA positive strains were subjected to cagPAI typing through 11 sets of primers spanning whole cagPAI locus, which were designed by Yadegar et al. (29). PCR reaction mixture of 50 μL was used, containing 1X PCR buffer, 1.5 mM MgCl2, 200 mM of each dNTPs, 1 pmol of each primer, 1 μL of genomic DNA (approximately 150 ng), and 0.05 U/mL of SuperTaq™ DNA polymerase (HT Biotechnology Ltd., Cambridge, UK). cagPAI typing was done under the following conditions: initial denaturation at 94°C for 4 minutes, 30 cycles of denaturation at 94°C for 1 minutes, annealing at the indicated temperature for each reaction for 45 seconds, extension at 72°C for 1 min/kb, and then the final extension at 72°C for 10 minutes. While lack of ≥ 1 cagPAI locus defined partial cagPAI in the studied strains, the cagPAI was defined as intact if all the selected gene loci were present. Complete deletion of cagPAI in the cagA negative strains were determined using Luni1 and R5280 primers for cag empty site as described before (30).
2.3. RNA Extraction and cDNA Synthesis
Total RNA of the strains were extracted from the freshly grown bacteria in BHI broth medium containing the selective supplement and were incubated at 37°C for 3 days at microaerophilic conditions to obtain an optical density of 0.2 at 600 nm (~3 × 108 bacteria/mL). Accordingly, pellets of the cultures were prepared after centrifugation (12000 g, 5 minutes) followed by washing with sterile phosphate buffered saline (PBS, pH 8). Total RNA was extracted using RNeasy mini kit (Qiagen, USA), according to the manufacturer’s instructors. The RNA concentration was measured for each strain spectrophotometerically. All the RNA extracts were preserved at -70°C until use for cDNA synthesis. Reverse transcription was performed with Titan One Tube RT-PCR kit (Sigma-Aldrich) using same cagA primer, according to the manufacture’s protocol.
2.4. Cell culture and Infection Assays
To analyze expression of CagA in the human gastric cancer cell line, AGS (ATCC CRL-1739TM, IBRC, Tehran, Iran) was incubated in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, GrandIsland, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY, USA), 1% non-essential amino acid (Gibco, Grand Island, NY, USA), 100 U mL-1 of penicillin, and 100 μg mL-1 of streptomycin (Gibco, Grand Island, NY, USA) at 37°C in a humidified incubator (Memmert, Dusseldorf, Germany) containing 5% CO2. H. pylori strains cagA+ and cagA- were re-suspended in DMEM and added to 2 × 106 AGS cells at a multiplicity of infection (MOI) of 100. After incubation in a 5% CO2/95% air incubator for the required length of time, AGS cells were washed once with PBS (Sigma-Aldrich) to remove non-adherent bacteria. For imoniassy analyses, AGS cells with attached bacteria were collected by Rippa buffer (Sigma-Aldrich), according to the manufacture’s protocol.
2.5. Detection of CagA
Expression of CagA was also characterized at protein level. The freshly prepared bacterial pellets were washed with PBS and centrifuged at 4000 g for 15 minutes. The pellets were re-suspended in lysis buffer (PBS containing lysozyme, 10 mg/mL, EDTA, 2 mM, DNase I, 10 μg/mL, and protease inhibitor cocktail table; Roche, Germany). Sonication of the treated bacteria was done after incubation in 37°C for 30 minutes (Ultrasonic UP 200 H, 100 Watt on ice for 2 minutes). To separate soluble proteins from the bacterial cell lysates, the sonicated solution was centrifuged at 12000 g for 20 minutes at 4°C. Supernatant and pellet of each lysate was collected after addition of urea (9 M) and dithiothreitol (DTT, 70 mM) and stored at -70°C until use for immunoblot analysis. Dot blotting was performed to screen presence of CagA in the protein extracts. Briefly after blocking with bovine serum albumin (5%), polyvinylidene fluoride membrane was inoculated with 15 µL of the crude protein extracts (3 separate 5 µL inoculations) from cagA positive and negative H. pylori strains. The blots were washed 3 times with TBS-T buffer (0.05%, Tris-buffered saline with Tween 20) and incubated with mouse anti-CagA primary antibody (1:1000 dilutions) for 24 hours at 4°C (Abcam, USA). The HRP-conjugated secondary antibody was added to the blots at 1:5000 dilutions (Razi Biotech, Iran) after 4 washing steps, and incubated for 1 h at same conditions. The presence of CagA was determined after washing of the membrane and addition of TMB solution (3,3’,5,5’-tetramethylbenzidine, 10 mg/mL). To confirm our results, immunoblotting was performed. Accordingly, 50 µL of the protein samples from each strain were subjected to electrophoresis in 12% sodium dodecyl sulfate (SDS)- polyacrylamide gels (30 V, overnight at room temperature). Protein bands were then transferred to a polyvinylidene fluoride membrane in a transfer buffer (25 mMTris, 192 mM glycine, pH 8.3) using Mini-Trans-Blot electrophoretic transfer cell (Roche, Germany). The presence of the specific band for CagA was determined through the same procedure that was described above. CagA expression was also analyzed for the strains encoding cagA mRNA in a cell culture model (AGS, ATCC CRL-1739TM, IBRC, Tehran, Iran). Accordingly, during the assay, confluent AGS cell line was incubated in antibiotic free Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% non-essential amino acid (Gibco, Grand Island, NY, USA), at 37°C in a humidified incubator (Memmert, Dusseldorf, Germany) containing 5% CO2. Two freshly grown cagA+H. pylori strains and 1 cagA-strain were suspended in DMEM and added to AGS cells at a multiplicity of infection (MOI) of 100. After incubation for 6 h at the same conditions, the total protein was extracted from the infected cell lines using the Rippa buffer (Sigma-Aldrich), according to the manufacture’s instruction. CagA expression was characterized by immunoblotting using same antibodies. All the assays were performed in 2 replicates.
4. Results
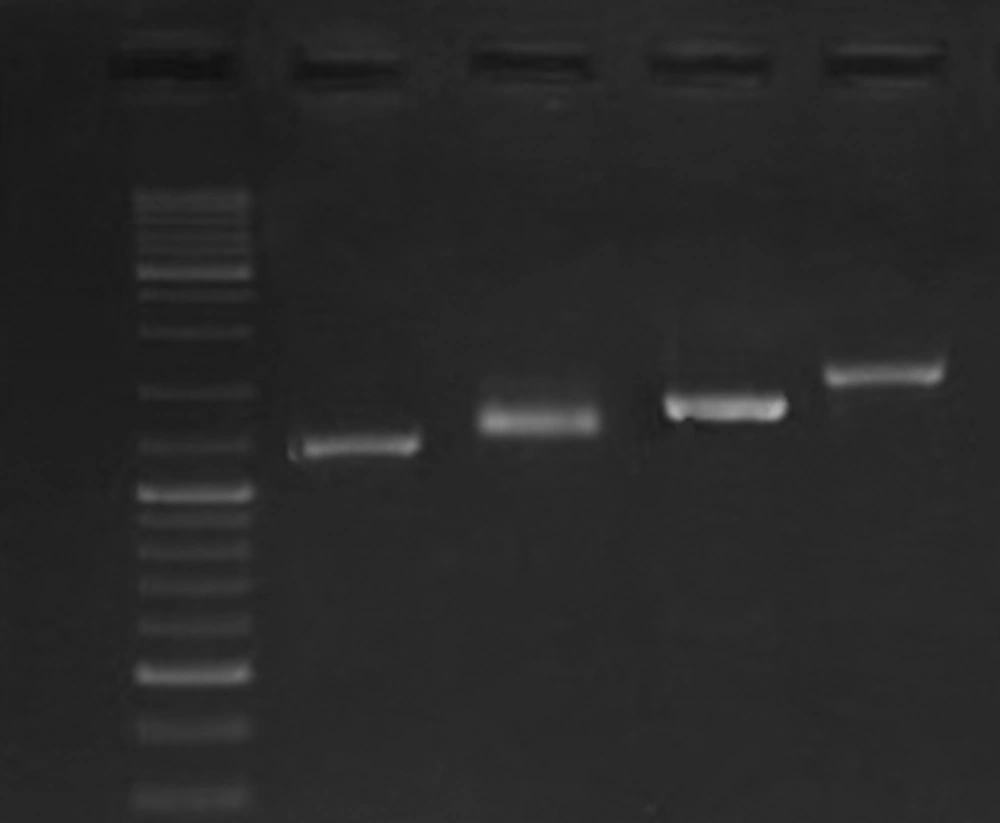
Identity of all the bacterial strains that were used in this study was confirmed by PCR results for glmM gene. PCR results showed the presence of cagA in 12 strains. One cagA negative strain was included in the study as a negative control for gene expression analysis. As was shown in Figure 1, based on diversity of the strains in cagA' variable region, different CagA EPIYA motifs were detected. Verification of the EPIYA types was done through sequence analysis of 1 amplicon that showed similar gel migration with the reference amplicon from H. pylori strain RIGLD-HC113 (Accession number KU516680). According to these results, we characterized the EPIYA types as follow: ABC, 7 strains (58.3%); ABCC, 4 strains (33.3%), and ABCCC, 1 strain (8.3%).
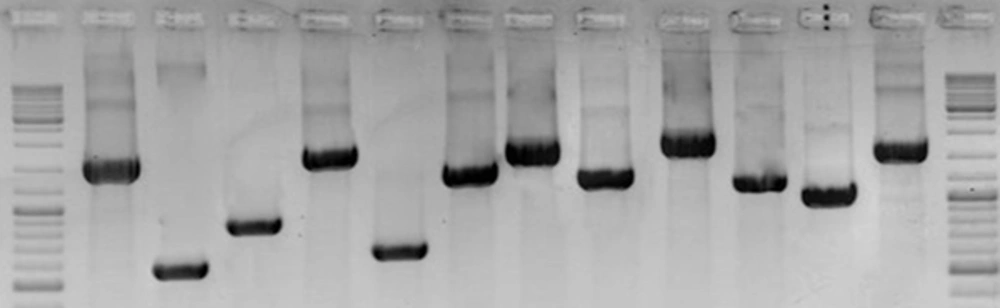
PCR analysis was performed on all strains using 11 pairs of spanning primers complementary to different cagPAI regions. The PCR amplifications yielded high-quality products with the expected sizes and reproducible results (Figure 2). The data presented in Table 1 show diversity of cagPAI genes and specified internal deletions (D1 to D5) among the strains with different EPIYA motifs. In total, 7 genotypes of cagPAI were determined in these strains. Intact cagPAI was detected among 4 strains (33%). Analysis of the results showed deletions D2 (1/12, 8.3%) and D3 (4/12, 33.3%), and occurrence of mutation at loci cagX-cagW (3/12, 25%), cagC-cagA (2/12, 16%), cagL-cagH (3/12, 25%), cagE-cagC (3/12, 25%), cagW-cagT (2/12, 16 %) in these strains.
Lane 1 and 14, GeneRuler DNA Ladder Mix (Thermo Scientific); Lane 2, cagζ-cagδ (Wild (intact): 1092 bp); Lane 3, D2 (525 bp); Lane 4, D3 (750 bp); Lane 5: cagQ-cagM (1721bp); Lane 6, cagp-cagM (601 bp); Lane 7, cagδ-cagγ (1417 bp); Lane 8: cagα-cagY (1818 bp); Lane 9, cagX-cagW (1306 bp); Lane 10: cagW-cagT (228 bp); Lane 11, cagL-cagH (1329 bp); Lane 12: cagE-cagC (1083 bp); Lane 13: cagC-cagA (1917 bp).
| H. pyloricagPAI Genotype | ID of Strains (Pathological Findings) | CagA Expression | CagA EPIYA Type |
|---|---|---|---|
| Intact cagPAIa | HC-113 (IM) | Positive | ABC |
| HC-168 (SAG) | Negative | ABC | |
| HC-180 (CAG) | Negative | ABCC | |
| HC-136 (SAG) | Negative | ABCC | |
| cagζ-cagδ (W)/cagα-cagY-/ cagX-cagW-/ cagC-cagA- | HC-176 | Negative | ABC |
| HC-174 (Moderate active chronic gastritis) | Negative | ABC | |
| cagζ-cagδ (D2)/cag -cag-/cagW-cagT-/cagL-cagH-/cagE-cagC- | OC-180 (Moderate active chronic gastritis) | Negative | ABC |
| cagζ-cagδ (D3)/cagX-cagW- | OC-179 (Moderate active chronic gastritis) | Negative | A/B |
| HC139 | Negative | ABCC | |
| cagζ-cagδ (D3)/cagQ (rev. ori.)+/cagQ-cagA-b | OC-287 | Negative | ABC |
| cagζ-cagδ (D3)/cagL-cagH-/cagE-cagC- | OC-217 (chronic active gastritis) | Negative | ABC |
| cagζ-cagδ (D3)/cagQ-cagA+c | OC-149 (chronic active gastritis) | Positive | ABCCC |
aIntact cagPAI: H. pylori strains with complete cagPAIgene segments.
bcagQ-cagA-: H. pylori strains with mutation at all genetic segments denoted between cagQ-cagM and cagC–cagA.
ccagQ-cagA+: H. pylori strains without mutation at all genetic segments denoted between cagQ-cagM and cagC-cagA. IM, intestinal metaplasia; SAG, severe active gastritis; CAG, chronic active gastritis.
We further assessed any correlation between EPIYA patterns and intactness of cagPAI locus. EPIYA types ABCC (3/12) and ABCCC (1/12) were observed dominantly among strains that showed most conserve cagPAI segments, while EPIYA type ABC (7/12) and A-B/C (1/12) was characterized among the strains with mutations at 2 or higher loci (Table 1).
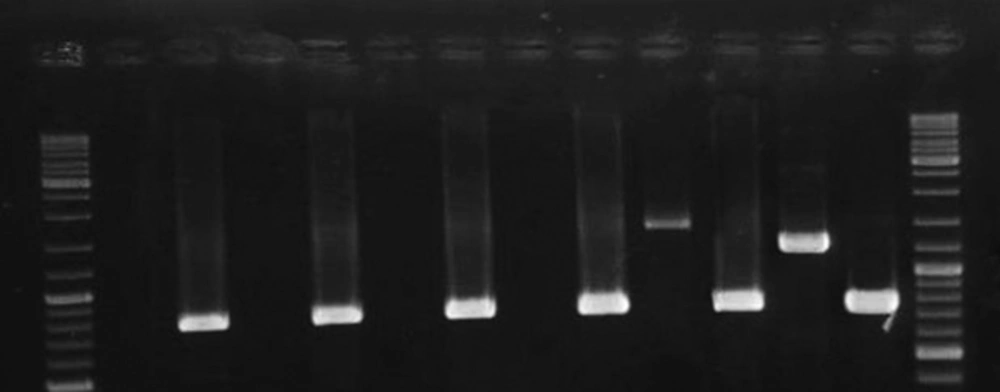
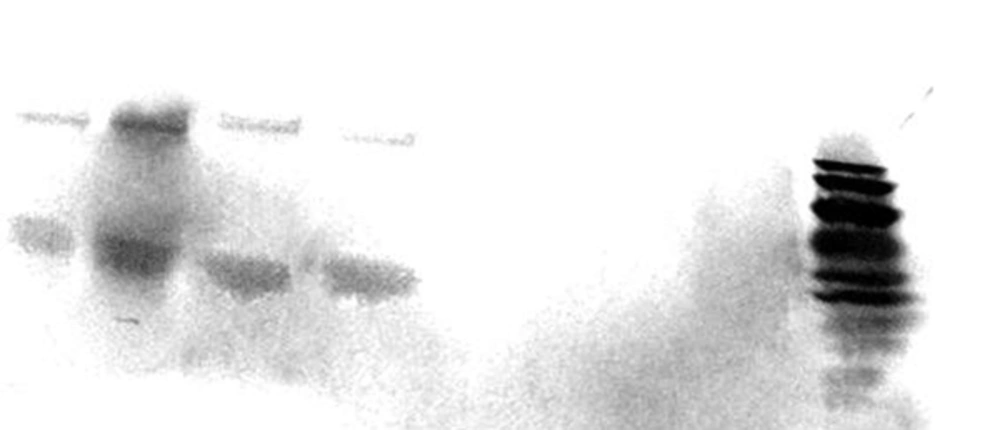
As was shown in Figure 3, cagA transcription was confirmed among 2 out of 6 H. pylori strains presented the most intact cagPAI genotypes. The 2 strains belonged to those carried intact cagPAI (Strain HC113, EPIYA-ABC) and 1 strain with D3 deletion (Strain OC149, EPIYA-ABCCC). Lack of cagA expression was detected in 3 strains with intact cagPAI, and one strain with mutation at cagX-cagW and cagζ-cagδ (D3) loci (Table 1). CagA expression was confirmed for these 2 strains by both immunoblotting and dot blotting methods and also in cell culture plates (Figure 4).
Lane 1 and 14, GeneRuler DNA Ladder Mix (Thermo Scientific, USA); Lanes 2, 4, 6, 8, non-coding cagAstrains (Strains OC217, HC168, HC136, HC180, respectively); Lanes 10 and 12, cagA coding strains (Strains OC149 and HC113, respectively). mRNA expression of 16srRNA was observed for all the strains; Lanes 3, 5, 7, 9, 11, and 13 represents the RT-PCR product for strains OC217, HC168, HC136, HC180, OC149 and HC113, respectively. Size variation of products for cagA is related to the nucleotide sequence diversity in these strains.
Lanes 1 - 4 represented CagA (complete and truncated form) protein in lysates of grown H. pylori strains in Brucella broth (Lanes 1 and 3) and total protein extracts of infected cell line (Lanes 2 and 4) after 6 h incubation. MOI of 100 was used in cell culture experiments; Lane 8, PageRuler™ Prestained Protein Ladder (10 - 170 kDa, Thermoscientific, USA); Lane 5, infectedAGS cell with cagA negative H. pylori strain (Strain OC236); Lane 6, Lysate of grown H. pylori strain OC236 in Brucella broth; Lane 7, Protein extracts of non-infected AGS cell line. Size variation for lanes 3- 4 are related to CagA EPIYA motif ABCCC.
5. Discussion
H. pylori has several housekeeping and virulence genes that support its persistent colonization and pathogenesis in the harsh gastric environment for decades. Pathogenesis of H. pylori is depended on diversity of these genes and function of their products in the host. CagPAI is one of the main H. pylori virulence markers that assumes as a mediator of inflammatory response and carcinogenesis through delivery of CagA into host cells (13, 31-33). While diversity of this structural component was described by some recent studies, its correlation with disease outcome is not yet clear. Our results showed different CagA EPIYA motifs (ABC, ABCC, ABCCC) in H. pylori strains isolated from the patients who suffered from mild gastritis, sever active gastritis, intestinal metaplasia, and gastric cancer. The existence of 7 different genotypes of cagPAI among the 12 studied strains confirmed a high rate of diversity in this genetic locus, which may correlate with pathogenicity of the strains. Ahmadzadeh et al. represented 13 different cagPAI genotyptes among 21 cagPAI positive H. pylori strains in Iran (34).This diversity is also high in different geographic populations (11, 35-37). Correlation between integrity of cagPAI, types of EPIYA motifs, and disease outcome was confirmed by several studies (24, 38-42). However, there are studies that reported contrary results (43-45). Variation in H. pylori genome from various areas and presence of polymorphisms in different gene loci of cagPAI could be main causes of the differences among the studies. In our study, composition of most important genetic loci of cagPAI was determined. The obtained results for genetic structure of cagPAI proposed the presence of functional deletions in different segments, including cagζ-δ, cagX-W, cagL-H, cagE-C, cagC-A, and cagW-T. Most of these segments encode critical structural proteins of CagPAI that are essential for interaction with host cells and delivery of H. pylori effector proteins. This finding could somewhat explain the variation in the clinical outcome of infected patients with distinct H. pylori strains. While there is some evidence regarding the effects of intact cagPAI on the severity of histopathological changes of the gastric tissue, ulceration, and carcinogenesis, establishment of this association needs further studies on different populations (39, 46-48).
Intact cagPAI could not solely guarantee correct function of type IV secretion system (T4SS) in H. pylori strains, since point mutations in cagPAI loci might cause nonsense mutations that disturb its activity. Selbach M. et al. showed that nonsense mutations in cagα (virB11), cagY (virB10), cagX (virB9), cagW (virB8), cagT (virB7), and cagE (virB4) could prevent CagA translocation, IL-8 induction, and formation of scattering phenotype in gastric epithelial cells (21). Necessity of functional CagY and CagL for delivery of CagA into host cells and secretion of inflammatory cytokine was established by Tegtmeyer N et al. (49). In our study, we found intact cagPAI in the 5 strains, ∆3 was preset in 1 of them. Gene expression analysis of cagA in these strains proposed a high frequency of nonsense mutation or mutation in promoter of this gene locus. In a study by John T. Loh et al., they revealed that strains expressing high levels of CagA were associated with more advanced precancerous lesions compared with those expressing low levels of CagA (50). The lower level of CagA expression was found to be in relation to the existence of a DNA motif upstream of the translational initiation site of CagA. In consistent to these results, high frequency of rearrangement in cagA was reported by Kauser F. et al.. They showed diversity in the cagA promoter as a main responsible factor affecting its expression in patients with severe pathology (17). The link between existence and higher expression levels of CagA and/or T4SS components with progression of gastric pathology and carcinogenesis was similarly depicted by Szkaradkiewicz A et al. (51). While our finding and those presented by other studies strongly indicate that intact cagPAI in strains with higher CagA expression level are more virulent, the expression of CagA and intactness of cagPAI may be unrelated, since there are some reports that showed inconsistency between integrity of cagPAI and expression of cagA (52).
Increase in EPIYA motifs and existence of some defined EPIYA types are associated with more strengthen intracellular signaling in the infected host cells (13, 38, 53-55). In another study, H. pylori strains with higher EPIYA type C-repeats showed higher frequency in the infected patients with gastric cancer (56). According to the previously determined EPIYA types of the studied strains in the previous study, our analysis showed no significant correlation between diversity of cagPAI and diversity in cagA EPIYA types. However, the presence of 2 or 3 EPIYA type C motifs was mainly observed in the stains with more conserved cagPAI loci, while strains that presented EPIYA type ABC were generally detected among the strains with partial cagPAI. This could presume ability of more virulent H. pylori strains for preservation of their integrity after the occurrence of the genetic recombination events. Effective interaction of CagPAI with host cell receptors may explain diversity of distinct H. pylori pathogenesis in these patients. Our results represented more conserved pathogenicity locus in severe gastritis patients, even among the strains with lower repeats of EPIYA motif. However, involvement of other H. pylori virulence factors or host factors also seems probable (25, 57-60).
4.1. Conclusion
In conclusion, our results showed a high frequency of partial cagPAI among the H. pylori strains in Iran. While all the selected strains were cagA positive, the presence of cagA didn’t predict the integrity of cagPAI or its expression. Therefore, assessment of all genetic loci seems to be necessary for detection of intact cagPAI in H. pylori strains. Lack of CagA expression in the cagA positive strains proposed high frequency of nonsense mutations in its coding sequence or the promoter region. The pathological findings proposed H. pylori strains with intact cagPAI and ability of CagA expression as more virulent strains than those with disruption at these loci.