1. Background
Acinetobacter baumannii has emerged worldwide as a major nosocomial pathogen and it is a great threat to hospitalized patients, particularly intensive care unit (ICU) patients (1, 2). Because of its ubiquitous nature, A. baumannii is isolated from hospital environments and clinical specimens. Multidrug resistance (MDR), extensively drug resistance (XDR), involvement in hospital outbreaks, and long-term persistence in hospital settings are the hallmarks associated with A. baumannii infections (1).
An unambiguous identification method is needed for A. baumannii isolates in order to identify the genetic relatedness and the population structure of A. baumannii strains, as well as to study the impact of public health measures on controlling the MDR infections. Until now, several typing methods have been developed in order to determine the genetic diversity, strains’ relatedness, and the epidemiology of A. baumannii. Among them, multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) are common methods being used in the molecular typing of A. baumannii (1, 3). MLST is a nucleotide sequence-based method used for unambiguous characterization of bacterial isolates. MLST provides an allelic profile or a sequence type for each isolate, which can be shared among the laboratories and throughout the world via the Internet. However, this method is expensive and inappropriate for the characterization of the isolates in the outbreaks (1, 4). In contrast, PFGE is considered the gold standard method in the molecular epidemiology of outbreaks. However, PFGE is labor-intensive, which takes several days to produce results and needs specific tools not available in all laboratories (1, 3). Therefore, in this study, a new approach of multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) was evaluated. MLVA is a faster and safer PCR-based typing method determining the copy numbers of tandem repeat units of VNTR loci. MLVA has some advantages including high resolution, data portability, and intra-laboratory reproducibility. It is also less expensive and less labor-intensive (5). MLVA is becoming a very popular typing method in the world. There have been many entries for articles in recent years; however, a few MLVA assays have achieved generally accepted standardization for the typing of bacterial pathogens (5).
A few MLVA schemas have been developed for A. baumannii typing, among which the MLVA-8Orsay scheme has been developed with high discriminatory power by Pourcel et al. (6).
2. Objectives
In the present study, the MLVA-8Orsay scheme was applied to evaluate its suitability for clustering and differentiating 89 carbapenem-resistant A. baumannii strains isolated from two hospitals in Tehran.
3. Methods
3.1. Bacterial Strains
A panel of 89 non-repetitive carbapenem-resistant A. baumannii isolates was selected from among well-characterized strains collected from various clinical specimens of hospitalized patients in a study conducted during 2010 - 2011 in two hospitals of Tehran city (7).
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed for all of the 89 strains of A. baumannii by the disk diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines. The test antibiotics were as follows: imipenem (10 µg), meropenem (10 µg), gentamicin (10 µg), ciprofloxacin (5 µg), amikacin (30 µg), trimethoprim-sulfamethoxazole (23.75 + 1.25 µg), cefepime (30 µg), cefotaxime (30 µg), aztreonam (30 µg), and ceftazidime (30 µg). Escherichia coli ATCC 25922 was used as a quality control strain. The minimum inhibitory concentrations (MICs) of imipenem and meropenem were determined using the agar dilution method (8). Strains resistant to at least three classes of antibiotics were considered MDR strains (9).
3.3. Molecular Detection of OXA Carbapenemase and Metallo-β-Lactamase
Four groups of OXA carbapenemase genes were detected by a multiplex PCR assay, including blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like genes (10). Ambler class B carbapenemase genes were detected by PCR and using specific primers for blaIMP and blaVIM (11).
3.4. MLVA Genotyping
Genomic DNA was prepared by the boiling method (12). The MLVA-8Orsay scheme was performed as described previously in a study by Pourcel et al. (6). Oligonucleotide primers targeting the flanking regions of the L-repeat VNTRs (Abaum1988, Abaum2240, Abaum3002, and Abaum3530) and S-repeat VNTRs (Abaum0826, Abaum0845, Abaum2396, and Abaum3468) were used for the amplification of A. baumannii genomic DNA by PCR. L (Larg)-repeat VNTR loci were the loci with a repeat unit size of above 9 bp, whereas small (S)-repeat VNTRs were the loci with a repeat unit size of up to 9 bp. The low level of polymorphism was observed in the L-repeat VNTR loci due to their slow evolving, whereas S-repeat VNTRs were very diverse. The clustering was achieved by combining L-repeat and S-repeat VNTRs (6). PCRs were performed in a 25 µL final volume containing 3 ng genomic DNA, × 1´ reaction buffer, 1.5 mM MgCl2, 1 U of Taq DNA polymerase, 200 µM of each deoxynucleoside triphosphate (dNTPs), and 20 pM of each primer. Amplification was performed with a Bio-Rad thermal cycler. PCR conditions were as follows: denaturation for 5 minutes at 94°C, followed by 36 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 50°C for Abaum0826 and Abaum0845 and at 55°C for the remainder loci, elongation for 50 seconds at 72°C, and a final elongation for 7 minutes at 72°C. After PCR, 3 µL of each PCR product was loaded on 2% regular agarose gel (Invitrogen, Carlsbad, CA, USA) for L-repeats and on 2% Metaphor agarose gel (FMC Bioproduct, Rockland, ME, USA) for S-repeats. Electrophoresis was performed in 25-cm wide gels and run at 4 V/cm in 0.5 × Tris-borate-EDTA buffers for about 5 hours. A 50-bp ladder (Bio-Rad) was used as a DNA size marker for L-repeat and S-repeat VNTRs. The gels were stained with 0.5 µg/mL of ethidium bromide for 1 hour and then photographed under UV illumination. The size of the amplicon was determined using the BioDocAnalyze (BDA) software. The length of repeat, the number of repetitions, and deduced size of the flanking regions were analyzed (https://tandem.bu.edu/trf/trf.html) (13, 14). Clusters analysis of the MLVA typing data was performed by MLVA Plus online tool. The polymorphism index of an individual locus or combined VNTR loci and confidence intervals (CIs) were calculated using the Hunter-Gaston diversity index (HGDI) by V-DICE software (Health Protection Agency, London, UK; http://www.hpa-bioinfotools.org.uk/cgi-bin/DICI/DICI.pl) (15).
4. Results
4.1. Antimicrobial Susceptibility Testing
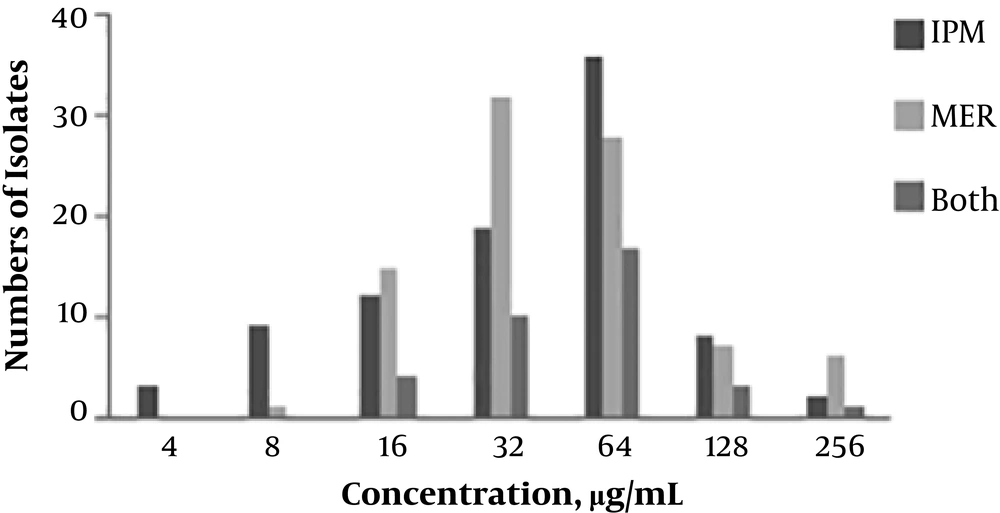
Antibiotic susceptibility testing revealed that 80.8% (72 out of 89) of the isolates were MDR. About 87.6% of the isolates were resistant to cefepime (78 out of 89), 61.7% to amikacin (55 out of 89), 80.8% to gentamicin (72 out of 89), 92.1% to trimethoprim/sulfamethoxazole (82 out of 89), and 100% to ceftazidime, aztreonam, imipenem, meropenem, cefotaxime, and ciprofloxacin (Table 1). The distribution of MICs for imipenem and meropenem in 89 A. baumannii isolates is shown in Figure 1. High-level resistance (with MICs ≥ 128 µg/mL) to imipenem, meropenem, and to both antibiotics was seen in 11.2, 14.6, and 4.4% of the A. baumannii isolates, respectively.
| IMP | MER | CPM | CTX | CAZ | ATM | GM | CIP | AN | STX | |
|---|---|---|---|---|---|---|---|---|---|---|
| Trachea, N = 45 | 100 | 100 | 86.6 | 100 | 100 | 100 | 91.1 | 100 | 62.2 | 95.5 |
| Wound, N = 24 | 100 | 100 | 83.3 | 100 | 100 | 100 | 66.6 | 100 | 35.5 | 91.6 |
| Others, N = 20 | 100 | 100 | 95 | 100 | 100 | 100 | 70 | 100 | 24.4 | 85 |
| Total, N = 89 | 100 | 100 | 87.6 | 100 | 100 | 100 | 80.8 | 100 | 61.7 | 92.1 |
Abbreviations: AN, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CPM, cefepime; CTX, cefotaxime; GM, gentamicin; IMP, imipenem; MER, meropenem; STX, trimethoprim-sulfamethoxazole.
4.2. Molecular Detection of Carbapenemase
The most prevalent carbapenemase was the blaOXA-23-like gene (83/89, 93.2%), followed by the blaVIM (15/89, 16.8%), blaOXA-24-like (5/89, 5.6%), blaIMP (5/89, 5.6%), and blaOXA-58-like (1/89, 1.1%) genes. All blaVIM gene-positive strains also carried the blaOXA-23-like gene. Two isolates carried three carbapenemase determinants, one isolate carried blaOXA-23-like, blaOXA-58-like, and blaVIM genes, and other isolates carried a combination of blaOXA-23-like, blaOXA-24-like, and blaVIM genes. Among six blaOXA-23-like gene-negative strains, one isolate was positive for the blaIMP gene, and five isolates were positive for the blaOXA-24-like gene. There was no relationship between high-level MIC and co-carriage of carbapenemase genes.
4.3. MLVA Genotyping
The occurrence of selected VNTR loci was studied in 89 A. baumannii strains. All L-repeat and S-repeat VNTR loci yielded amplification products ranging from 154 bp to 826 bp for most of the isolates (Table 2). In one of the isolates, a half-size allelic variant was observed at locus Abaum2240. In 10 isolates, locus Abaum1998 yielded an ambiguous amplification pattern with two bands; these uncommon patterns were defined as “a”. The amplification failure was observed in the isolates at Abaum3468 (22.4%), Abaum3530 (5.6%), Abaum2240 (6.7%), Abaum1998 (6.7%), Abaum0826 (7.8%), Abaum2396 (7.8%), and Abaum3002 (4.4%). As shown in Table 2, the highest diversity was observed at S-repeat VNTRs, resulting in HGDI values ranging from 0.89 (Abaum3468) to 0.96 (Abaum0845). The locus Abaum0845 showed a wide variability and generated alleles with high diversity in repeat numbers, from one repeat up to 64 repeats; therefore, due to the high diversity associated with Abaum0845 in this study and thus no further discriminatory power by adding Abaum0845, the Abaum0845 locus was excluded from the MLVA scheme. Therefore, the isolates were analyzed by seven loci (MLVA-7). The isolates (157T, 85T, 73T, 67T, 58T, and 49T) with amplification failures in more than three loci were excluded from further analysis.
| Locus | Repeat Size (bp) | HGDI | CIa | Kb | No. of Most Frequent Repeats (%) | No. of Amplification Failures (%) |
|---|---|---|---|---|---|---|
| Abaum-1998 | 26 | 0.764 | 0.750 - 0.779 | 11 | 9 (34.8) | 6 (6.7) |
| Abaum-3002 | 57 | 0.392 | 0.361 - 0.424 | 5 | 7 (76.4) | 4 (4.4) |
| Abaum-3530 | 60 | 0.499 | 0.473 - 0.534 | 6 | 6 (67.4) | 5 (5.6) |
| Abaum-2240 | 99 | 0.549 | 0.523 - 0.576 | 7 | 2 (62.9) | 6 (6.7) |
| Abaum-0826 | 9 | 0.927 | 0.922 - 0.932 | 23 | 19 (12.3), 20 (12.3) | 7 (7.8) |
| Abaum-2396 | 6 | 0.933 | 0.956 - 0.961 | 29 | 21 (14.6) | 7 (7.8) |
| Abaum-3468 | 6 | 0.898 | 0.888 - 0.908 | 27 | 17 (17.9) | 20 (22.4) |
| Abaum-0845 | 7 | 0.963 | 0.949 - 0.977 | 34 | 16 (7.8), 17 (7.8) | 5 (5.6) |
Abbreviation: HGDI, Hunter-Gaston diversity index.
a Precision of the diversity index expressed as 95% upper and lower boundaries.
b Number of different repeats present at this locus in this sample set.
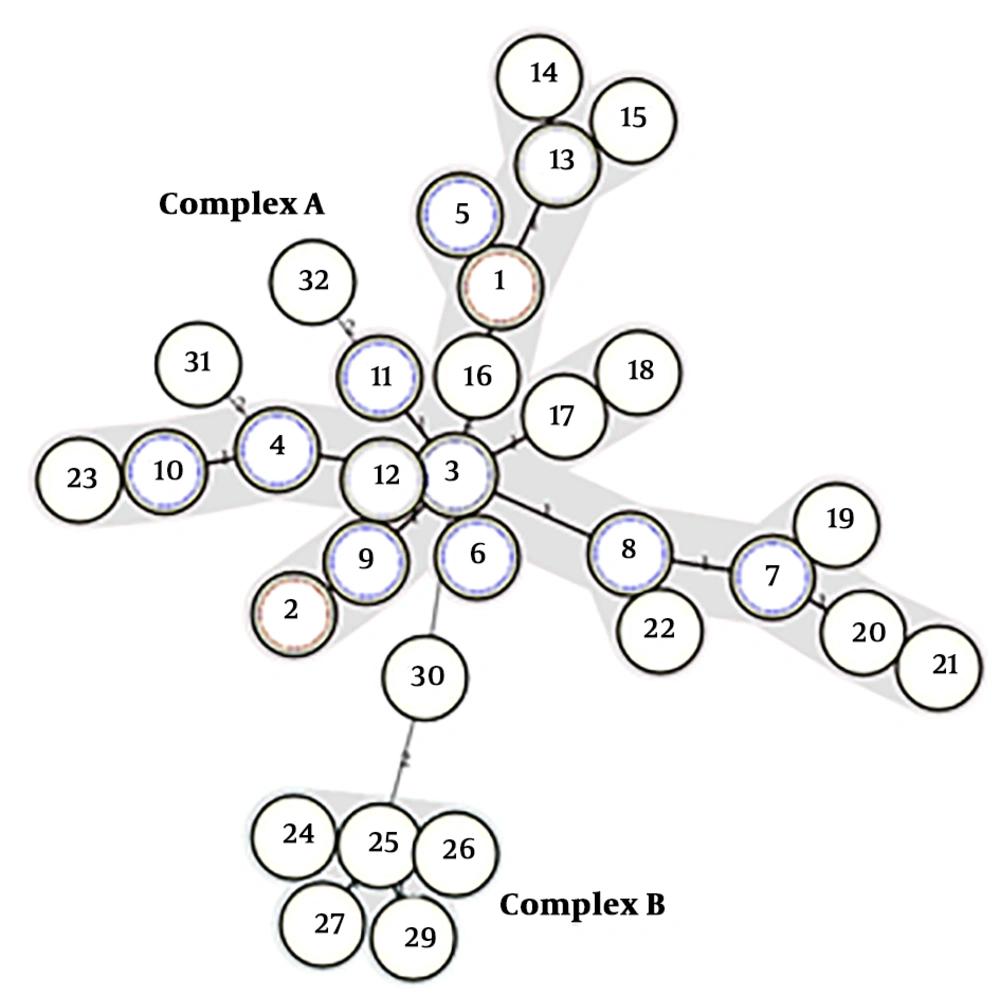
The 83 isolates were distinguished by 25 different MLVA types, as represented by individual allelic profiles of L-repeat VNTRs (Figure 2). MLVA type 1 and 2 were the most prevalent types containing 19 and 12 members with identical allelic profiles of L-repeat VNTRs (allelic profiles of 6-7-2-3 and 7-7-3-9, respectively), followed by MLVA type 3 with seven members, MLVA type 4 with five members, MLVA type 5, 6, and 7 with four members, MLVA type 8, 9, 10, 11, and 12 with three members, and MLVA type 12 and 13 with two members. The rest of the MLVA types included only one isolate. As shown in Figure 2, with a cut-off value of 75% corresponding to the existence of identically sized alleles for at least three L-repeat VNTRs, one clonal complex and three singletons were generated.
Cluster analysis of A. baumannii isolates based on four MLVA L-markers. A, MLVA dendrogram based on profiles of four L-markers for the 83 A. baumannii isolates. B, Minimum spanning tree representation of the L-locus clustering for the 83 A. baumannii isolates with one locus difference. Each circle indicates a unique MLVA type. The numbers within the circles denote strains belonging to the same MLVA type. Solid lines connect single locus variant MLVA types and the dashed lines indicate two loci variant MLVA types. The clonal complex was marked by a grey halo.
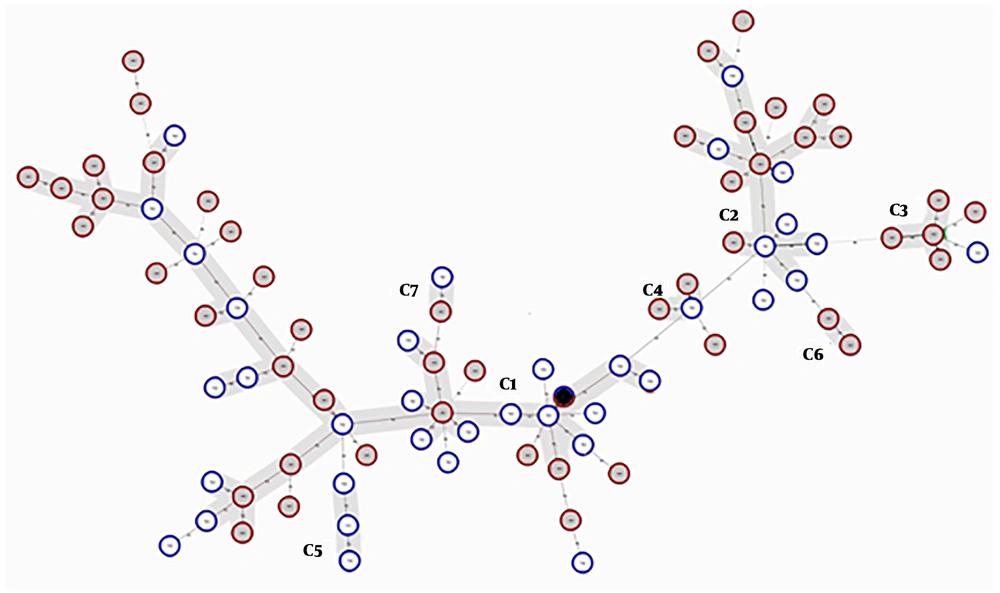
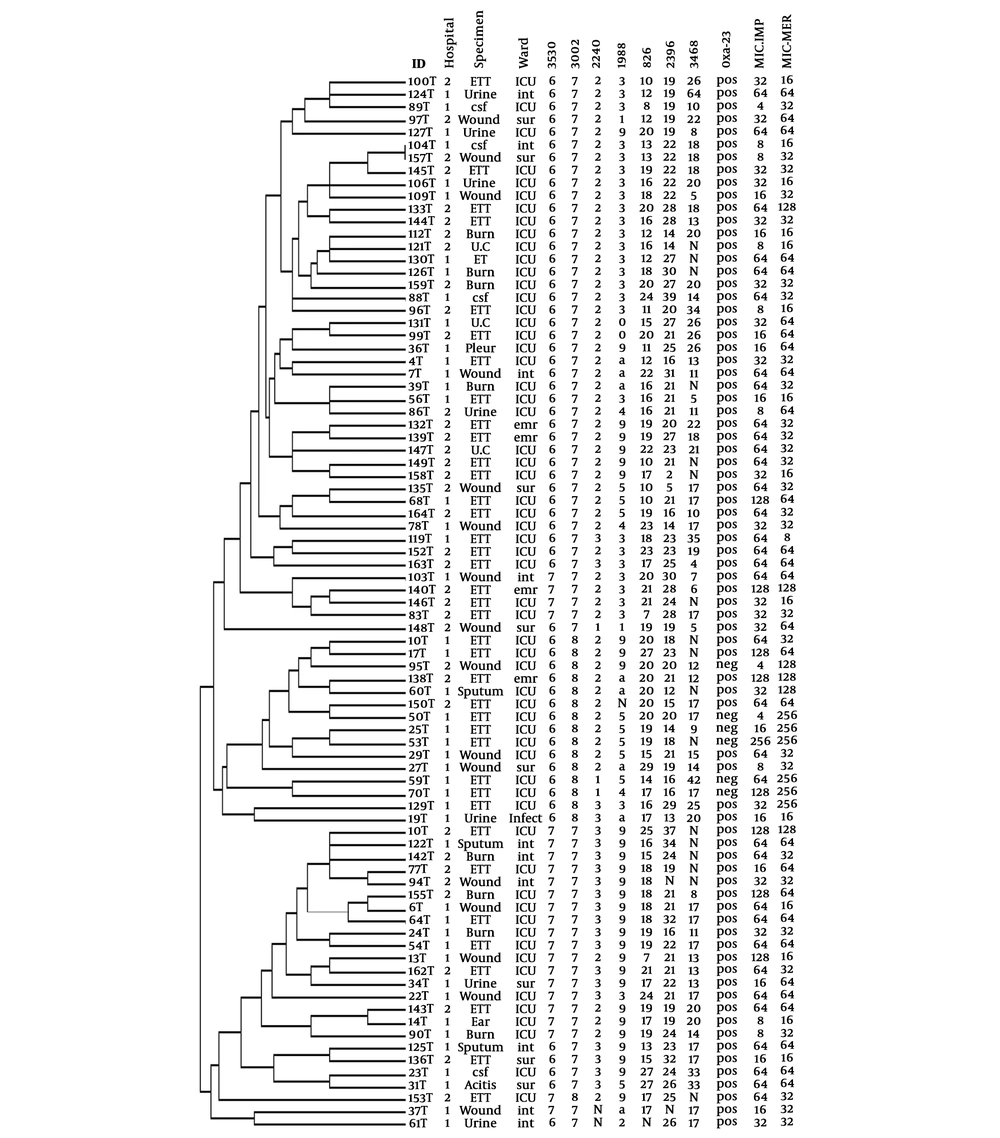
Finally, 82 different MLVA types with individual allelic profiles for 83 isolates were defined with the MLVA-7 assay. A combined HGDI value of 0.95 was achieved, consistent with the high level of polymorphism of these VNTR loci. Figure 3 shows the minimum spanning tree (MST) clustering of A. baumannii isolates. Due to the existence of identically sized alleles for at least five VNTRs, six clonal complexes (two isolates or more) and 17 singletons were observed. As shown in Figure 3, the clonal complex 1 (CC1) with 34 isolates and CC2 with 16 isolates were the main clusters, and the rest of the CCs included two or three isolates. The OXA-23 carrying isolates were dispersed in all of the six clonal complexes; four OXA-23 negative isolates were distributed to CC1, and two OXA-23 negative isolates were singletons. The other carbapenemase gene-carrying isolates were distributed to all of the CCs and/or singletons. CC5 contained only the isolates from hospital 1 and CC6 contained only the isolates from hospital 2, and the rest of the CCs contained isolates from both hospitals. MLVA-7 dendrogram and characteristics of A. baumannii strains are shown in Figure 4.
Minimum spanning tree representation of MLVA-7 clustering for the 83 A. baumannii isolates. Each circle indicates a unique MLVA type. MLVA complexes were defined assuming a 71% genetic similarity cut-off, corresponding to the existence of identically sized alleles for at least five VNTR markers. A black circle included two isolates from hospitals 1 and 2 with identical allelic profiles and six complexes were marked by grey halos. Solid lines connect two loci variant MLVA types and the dashed lines indicate three loci variant MLVA types.
MLVA-7 allelic profiles and characteristics of A. baumannii strains. The categorical coefficient and UPGMA algorithm were used for the generation of the dendrogram. ETT; endotracheal tube, ICU; intensive care unit, sur; surgery, emr; emergency, int; internal medicine, infect; infectious, N; no amplification, a; two bands pattern, MIC; µg/mL
5. Discussion
A. baumannii is one of the most common pathogens associated with hospital-acquired infections, which is almost resistant to all available antibiotics (2). To some extent, the success of A. baumannii in adaptation to the hospital environment has been attributed to its ability in acquiring resistance to almost all antibiotics, including carbapenems (4). Carbapenems are used as the last resort for the treatment of MDR strains of A. baumannii; however, the acquired carbapenem-hydrolyzing OXA-type class D β-lactamases (CHDLs) and metallo β-lactamase producing strains have been reported worldwide (1, 4). The intrinsic blaOXA-51-like gene family, the acquired blaOXA-23-like, blaOXA-24/40-like, and blaOXA-58-like gene families are considered the four main families of CHDL genes that have been reported for A. baumannii (1). The emergence and rapid expansion of CHDL gene-carrying strains have been found to be associated with hospital-acquired infections and outbreaks in hospitals, particularly in intensive care units (ICUs) (4). In the present study, it was found that blaOXA-23-like gene (93.2%) was the most prevalent gene in carbapenem-resistant isolates from the two hospitals in Tehran. The emergence of the blaOXA-23-like gene in clinical isolates of A. baumannii has been reported in other studies conducted in Iran (7, 16). Clonal outbreaks of blaOXA-23-like gene-carrying A. baumannii strains have also been reported in other studies conducted in France and Brazil (17, 18). In this study, one (mainly blaOXA-23-like gene), two, or three carbapenemase genes were detected in all of the carbapenem-resistant isolates. In addition, all the carbapenemase gene-carrying isolates were resistant to imipenem and/or to meropenem. However, there was no relationship between high-level MICs and co-carriage of carbapenemase genes. While OXA carbapenemase and MBLs are considered the main resistance mechanisms in A. baumannii, the co-existence of other resistance mechanisms such as increased expression of efflux pumps, loss of porins and other carbapenemase genes such as KPC carbapenemase, NDM, and SIM metallo-beta-lactamase genes can affect the levels of resistance (1, 18, 19). Thus, the presence of the aforementioned resistance mechanisms probably has affected the level of MICs in our isolates. In addition, the expression of carbapenemase genes can be affected by the presence of insertion elements in the upstream of the OXA gene (20).
MLVA has been developed for typing of various microbial species including Escherichia coli O157, Salmonella enterica, Pseudomonas aeruginosa, and A. baumannii (21-23). In the current study, MLVA-8Orsay scheme was performed in order to trace the epidemiology of the carbapenem-resistant A. baumannii isolates and for bacterial differentiation at the strain level. The MLVA-8Orsay scheme is based on eight VNTR markers, four of which are L-repeat VNTR markers (repeat units of 26 to 99 bp) with moderately diverse repeats, showing low levels of polymorphism, and the rest of the markers are highly diverse S-repeat VNTR markers (repeat units of 6 to 9 bp), characterized by a large number of alleles with high levels of variability (6). Among S-repeat VNTR markers, Abaum0845 was excluded from the MLVA scheme in the current study due to the high allelic diversity of this locus in the isolates tested. In the current study, the presence of Abaum0845 in the MLVA scheme caused a great diversity so that it was not possible to cluster the isolates, making it difficult to interpret the obtained results. However, when the Abaum0845 locus was excluded, the MLVA-7 scheme generated six complexes with a 71.4% genetic similarity cut-off. Abaum0845 was also absent in all A. baumannii isolates tested in another study conducted in Iran (16). In addition, variability associated with Abaum0845 was reported in another study conducted in Spain (24). It is possible that this locus is a rapidly evolving marker. In addition, six isolates showed no products at more than three loci including L-repeat and S-repeat VNTR markers, possibly due to the differences in the genetic structure of these isolates at the corresponding loci. These six isolates were also excluded from further analysis by MLVA-7 scheme and other VNTR markers or typing methods are needed for their genotyping. The amplification failure at various VNTR markers was shown in the registered isolates of A. baumannii in the MLVA bank for Acinetobacter genotyping (http://microbesgenotyping.i2bc.paris-saclay.fr/databases/view/202). The website of MLVA bank 2016.V1.3.5 is hosted by the University of Paris-Saclay, Orsay, France.
A total number of 83 A. baumannii isolates were typed into 25 different MLVA types using four L-repeat loci. Using a cut-off value of 75% corresponding to the existence of identically sized alleles for at least three L-repeat VNTRs, one clonal complex was achieved, showing the suitability of these markers for studying the phylogenetic relationships among the strains. Interestingly, among L-loci, Abaum1998 yielded an ambiguous amplification pattern with two bands in 10 isolates. In a study by Hu et al., two different repeat units were obtained for locus Abaum1998 in all the tested isolates; thus, this locus was excluded from their MLVA scheme (23). However, in this study, an uncommon pattern was defined by “a” for 10 isolates and analyzed as an individual pattern.
Pourcel et al. proposed the criteria for the assignment of the strains to MLVA complexes as (i) identical L-repeat VNTR profiles and (ii) a maximum of three differences of no more than two repeats at S-repeat VNTR loci (6). The intrinsic diversity of L-repeat VNTRs was higher in the isolates of the current study than in the isolates used in Pourcel et al. study; therefore, our MLVA complexes were not grouped with identical L-repeat VNTR profiles. In addition, in this study, the difference was observed in more than two repeats at S-repeat VNTR loci; thus, due to the diversity associated with MLVA-7 markers, applying the Pourcel et al. criteria was impossible. However, the MLVA complexes in this study were defined assuming a 71% genetic similarity cut-off, corresponding to the existence of identically sized alleles for at least five VNTR markers (Figure 3). By combining seven MLVA markers, an excellent discriminatory power was achieved (HGDI = 0.99), which is suitable for diseases surveillance and/or outbreaks investigation.
The 77 OXA-23 carrying isolates were distinguished by 76 MLVA-7 types within the six clonal complexes (Figure 4). Only did two OXA-23 gene-carrying isolates, i.e., 104T and 157T, share the same MLVA-7 allelic profile, which were isolated from hospital 1 and 2, respectively. The other families of carbapenemase gene-carrying isolates were also distributed to different clonal complexes with individual MLVA-types. No correlation was observed between MLVA-7 defined complexes and carbapenemase gene carriage and the origin of the isolates. Overall, the MLVA-7 analysis revealed that carbapenemase gene-carrying A. baumannii isolates in this study might be polyclonal (corresponding to 76 MLVA-7 types) and provided evidence for gene transfer among different MLVA-types in two hospitals in Tehran.
In conclusion, the main purpose of the study was to evaluate the MLVA-8Orsay scheme for typing of clinical A. baumannii isolates. We had some limitations with MLVA-8Orsay for typing the test isolates. First, Abaum0845 was excluded from the MLVA scheme because of its great allelic diversity observed in our isolates, making it difficult to cluster test isolates. Second, Abaum1998 yielded two bands in 10 isolates; these uncommon patterns were considered as distinct patterns. Third, nine isolates were excluded from further analysis due to amplification failure at more than three VNTR loci. Fourth, we did not apply Pourcel et al. criteria for the interpretation of the results and clustering the isolates due to the high diversity associated with MLVA-7 markers. However, the discriminatory power of MLVA-7 was found to be excellent and possibly useful and reliable in epidemiological investigations of A. baumannii. If MLVA-8Orsay was developed with new S-markers, it would be more helpful for better interpretation and clustering of the isolates.
This method is PCR-based, making it feasible in many laboratories. The results also provided evidence for the possible use of MLVA-7 scheme as an alternative or complementary method for typing of A. baumannii isolates.