1. Background
Escherichia coli strains are among the major causes of diarrhea particularly in infants in developing countries (1). The virulence factors associated with E. coli mediated diarrhea have been used for classification of strains as distinct diarrhoeagenic pathovar. Enteropathogenic Escherichia coli (EPEC) is one of the diarrhoeagenic pathovars that belongs to an attaching and effacing (A/E) lesion forming family of pathogens; this family includes enterohaemorrhagic E. coli (EHEC), rabbit diarrhoeagenic E. coli (RDEC), Escherichia albertii, and the murine pathogen Citrobacter rodentium (2). The A/E lesions are comprised of bacteria intimately bound to the host cell surface leading to damage of microvilli and pedestal formation. The pathogenicity island locus of enterocyte effacement (LEE) required for the production of these lesions. The core region of LEE is highly conserved, which encodes the type III secretion system, whereas a higher diversity observes among genes encoding effector proteins. EPEC strains can be classified into typical EPEC (tEPEC) and atypical EPEC (aEPEC) subgroups based on the presence or absence of the type IV-like bundle-forming pili (BFP) that encoded in a ca. 90-kb E. coli adherence factor plasmid (pEAF) (3). Besides LEE pathogenicity island (PAI), various PAIs have been found on chromosomes of EPEC strains, such as pathogenicity island OI-122.
OI-122 is a 23-kb pathogenicity island with three distinct modules in O157:H7 strain EDL 933. Module 1 carries pagC gene, module 2 carries sen, nleB and nleE genes and the third module contains efa1/lifA and efa2 genes, these modules are separated from each other by various IS elements (4). However, it was reported in various forms, complete with three distinct modules and partially with one or two modules (5). It has been shown that the presence of the OI-122 genes (i.e. efa1/lifA, sen, pagC, nleB, and nleE) in aEPEC is statistically linked with severe diseases and outbreaks (6). Other studies have found that OI-122 was more prevalent in tEPEC than in aEPEC and significantly associated with diarrhea in aEPEC strains (7, 8). In our previous study, we investigated the prevalence of the EPEC strain in clinical E. coli isolates from 363 stool samples of children from 2011 to 2013. Our results showed the presence of the EPEC strains in 42 of the isolates (9). The present study was designed to characterize the pathogenicity O island 122 (OI-122) among EPEC strains and to investigate the epidemiological relationship using MLVA.
2. Objectives
The aim of this study was to investigate the distribution patterns of this genomic island in Iranian tEPEC and aEPEC strains by detecting OI-122 genes and serogrouping, multi locus VNTR (MLVA) typing, and phylogenetic grouping.
3. Methods
3.1. Bacterial Strains
Forty-two EPEC strains, defined as eae positive E. coli without stx1 and stx2 genes, were collected from stool samples from children with diarrhea (≤ 10 years old), in the study conducted during the period from 2011 to 2013 in the four pediatric hospitals in Tehran, Iran (9). The 16 typical (bfpA+) and 26 atypical (bfpA-) EPEC isolates were stored at -70°C until further analysis.
3.2. Detection of OI-122 Genes
All of the EPEC isolates were screened by PCR for the presence of OI-122 genes (i.e. efa/lifA, sen, pagC, and nleB) using primers described elsewhere (7-11). Genomic DNA of E. coli strain EDL933 was used as positive control (Table 1).
| Gene | Primer Sequence (5’ to 3’) | Size of Amplicon, bp | Temperature, °C |
|---|---|---|---|
| chuA | F: GAC GAA CCA ACG GTC AGG AT | 279 | 55°C |
| R: TGC CGC CAG TAC CAA AGA CA | |||
| yjaA | F: TGA AGT GTC AGG AGA CGC TG | 211 | 55°C |
| R: ATG GAG AAT GCG TTC CTC AAC | |||
| TspE4C2 | F: GAG TAA TGT CGG GGC ATT CA | 152 | 55°C |
| R: CGC GCC AAC AAA GTA TTA CG | |||
| efa/lifA | F: AGA ATG GAA GAT CAC ACC | 309 | 55°C |
| R: ATA ATG CCT TTC ATC CAC AC | |||
| nleB | F: GCT TTC ACC GAT AAG GAC AAC | 272 | 55°C |
| R: TCG CCA TCA ACA AAA ATA CC | |||
| pagC | F: ATG AGT GGT TCA ACA CTG | 520 | 54°C |
| R: CCA ACT CCA ACA GTA AAT CC | |||
| sen | F: GGA TGG AAC CAT ACC TGG | 550 | 58°C |
| R: CGC AAT CAA TTG CTA ATG C | |||
| ms06 | F: AAA CGG GAG AGC CGG TTA TT | 39 | 55°C |
| R: TGT TGG TAC AAC GGC TCC TG | |||
| ms07 | F: GTC AGT TCG CCC AGA CAC AG | 39 | 55°C |
| R: CGG TGT CAG CAA ATC CAG AG | |||
| ms09 | F: GTG CCA TCG GGC AAA ATT AG | 179 | 55°C |
| R: CCG ATA AGG GAG CAG GCT AGT | |||
| ms11 | F: GAA ACA GGC CCA GGC TAC AC | 96 | 55°C |
| R: CTG GCG CTG GTT ATG GGT AT | |||
| ms21 | F: GCT GAT GGC GAA GGA GAA GA | 141 | 55°C |
| R: GGG AGT ATG CGG TCA AAA GC | |||
| ms23 | F: GCT CCG CTG ATT GAC TCC TT | 375 | 55°C |
| R: AAC TGG CGG CGT TTA TCA AG | |||
| ms32 | F: GAG ATT GCC GAA GTG TTG C | 101 | 55°C |
| R: AAC TGG CGG CGT TTA TCA AG |
Primers Used in This Study for Detection of Phylogenetic Groups, the Virulence Genes and MLVA Typing
3.3. Phylogenetic Grouping
Phylogroup determination for EPEC isolate was accomplished using the previously described PCR methodology (12). Briefly, the chuA and yjaA genes and DNA fragment TSPE4.C2 amplified by PCR. The phylogenetic group A (negative for all three genes), B1 (TSPE4.C2 positive), B2 (both chuA and yjaA genes positive) and D (chuA positive) was detected based on presence or absence of the chuA and yjaA genes and DNA fragment TSPE4.C2.
3.4. Serogrouping
The slide agglutination test was performed using EPEC O-specific polyvalent antisera (Mast, UK) according to manufacturer’s instructions. The polyvalent antisera consist of three separated pools, able to react with the following serogroups: poly group 2 (O26, O55, O111, O119, and O126), poly group 3 (O86, O114, O125, O127 and O128) and poly group 4 (O44, O112, O124 and O142). The agglutinated isolates further tested by monovalent O antisera.
3.5. MLVA Genotyping
MLVA genotyping were performed with the primer for seven loci (ms06, ms07, ms09, ms11, ms21, ms23, and ms32) (13). Repeat units were imported into Microsoft Excel 2007. The minimum spanning tree (MST) was constructed with a categorical coefficient based on allelic profiles of the strains (www.MLVAplus.net). Clonal complexes (CCs) are defined as a group of allelic profiles in which every profile shares at least 5 loci in common with at least one other member of the group (14). Genetic diversity of each VNTR locus was determined by calculating the Simpson’s index of diversity (SID) using the V-DICE software (Health Protection Agency, London, UK; http://www.hpa-bioinfotools.org.uk/cgi-bin/DICI/DICI.pl). To evaluate congruence between MLVA CCs and the other typing methods, the adjusted Wallace coefficient (AW) was calculated (www.comparingpartitions.info).
3.6. Statistical Analysis
Fisher exact test was used for data analysis. A P value of < 0.01 was considered statistically significant.
4. Results
4.1. Phylogenetic Group Determination
The most common phylogenetic group among tEPEC and aEPEC strains were B1 (68.75% and 80.76%, respectively). 4.6% of the aEPEC strains belonged to phylogenetic group A, 7.6% strains belonged to phylogenetic group D and one isolate belonged to phylogenetic group B2. 25% of the tEPEC strains belonged to phylogenetic group D and one isolate belonged to phylogenetic group B2 (Table 2).
| Strain | efa/lifA | sen | nleB | pagC | Phylogroup | Serogroup | MLVA Type | Hospital |
|---|---|---|---|---|---|---|---|---|
| tEPEC Strains | ||||||||
| TMU29 | n | n | p | n | B1 | O128 | M1 | B |
| TMU53 | n | n | p | n | B1 | ONT | M2 | M |
| TMU75 | n | n | n | n | B1 | ONT | M3 | B |
| TMU112 | p | n | p | p | B1 | O127 | M4 | A |
| TMU135 | n | n | p | n | B1 | O127 | M5 | M |
| TMU145 | n | n | p | n | D | ONT | M6 | A |
| TMU155 | p | n | p | n | B1 | O128 | M7 | A |
| TMU157 | n | p | p | n | D | ONT | M8 | M |
| TMU164 | n | p | p | p | D | ONT | M9 | M |
| TMU168 | n | n | p | p | B1 | ONT | M10 | A |
| TMU172 | n | p | p | p | B2 | O111 | M11 | B |
| TMU192 | p | p | p | p | B1 | ONT | M12 | A |
| TMU226 | p | p | p | p | B1 | ONT | M13 | B |
| TMU337 | p | n | n | n | D | O44 | M14 | H |
| TMU346 | p | n | p | n | B1 | O111 | M15 | H |
| TMU391 | n | n | p | n | B1 | O111 | M16 | H |
| aEPEC Strains | ||||||||
| TMU16 | n | n | n | n | A | ONT | M17 | M |
| TMU19 | n | n | p | n | D | ONT | M18 | M |
| TMU45 | n | n | n | n | B1 | ONT | M19 | A |
| TMU49 | n | n | n | n | B1 | ONT | M20 | M |
| TMU81 | n | n | p | n | B1 | O111 | M21 | M |
| TMU93 | n | n | p | n | B1 | O55 | M22 | B |
| TMU106 | n | n | p | n | B1 | O128 | M23 | M |
| TMU211 | n | n | n | n | B1 | O128 | M23 | M |
| TMU120 | n | n | p | p | B1 | O55 | M24 | A |
| TMU130 | n | n | n | n | B1 | O111 | M25 | A |
| TMU132 | p | n | p | p | D | ONT | M26 | B |
| TMU180 | n | n | n | n | B1 | O128 | M27 | A |
| TMU188 | n | n | n | n | B1 | O128 | M28 | A |
| TMU194 | n | n | n | n | B1 | O111 | M29 | M |
| TMU202 | n | n | n | n | B1 | O44 | M30 | A |
| TMU243 | n | n | p | n | B1 | O128 | M31 | B |
| TMU252 | n | n | n | n | A | ONT | M32 | H |
| TMU261 | n | n | n | n | B1 | ONT | M33 | B |
| TMU264 | n | n | n | n | B1 | ONT | M34 | B |
| TMU268 | n | n | n | n | B1 | ONT | M35 | B |
| TMU278 | n | p | n | n | B2 | ONT | M36 | B |
| TMU329 | n | n | n | n | B1 | O55 | M37 | A |
| TMU353 | n | n | p | n | B1 | O127 | M38 | H |
| TMU362 | n | n | n | n | B1 | O55 | M39 | A |
| TMU369 | n | n | n | n | B1 | ONT | M40 | A |
| TMU380 | n | n | n | n | B1 | O55 | M41 | A |
Comparison of Phylogenetic Classification, OI-122 Genes, Serotype, MLVA Typing for Each of the 42 EPEC Strains Identified in a Study Among Children with Acute Diarrhea
4.2. Serogrouping
The most common serogroups among EPEC isolates were found to be the members of O128 (n = 7, 16.7%) and O111 (n = 6, 14.3%), followed by O55 (n = 5, 11.9%), O127 (n = 3, 7%), and O44 (n = 2, 4.8%). However, the majority of the isolates (n = 19, 45.2%) were untypeable (ONT) with the antisera used (Table 2).
4.3. MLVA Types
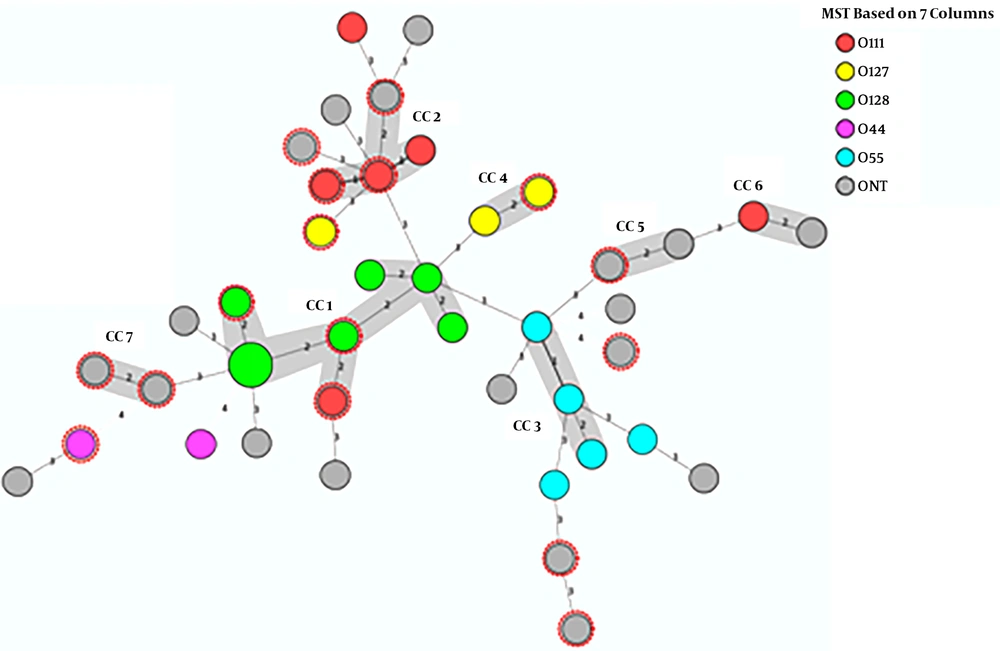
The MLVA identified 41 different allelic profiles (MLVA types) among 42 EPEC isolates. The Simpson's diversity of index was 0.975 that indicated the high discriminatory power for typing of EPEC isolates. An existence of identical size alleles for at least 5 VNTR markers, corresponding to the 71% genetic similarity cut-off was used to define MLVA clonal complexes (CCs) in this study. Therefore, 23 (54.8%) isolates were classified into 7 CCs, while the remaining isolates (45.2%) were assigned singletons. MLVA-CC1 was the largest lineage (clone) consisting of 7 MLVA types (Figure 1).
Minimum spanning tree (MST) based on MLVA data of EPEC isolates. The circles indicate the MLVA types and the size of the circles indicate the number of isolates. The number of loci that differ between two MLVA types is indicated on the lines connecting the MLVA types. The colours of the circles indicate the serogroups of the isolates and clonal complexes (CCs) are indicated by grey halos. The red dotted circles represent tEPEC isolates.
MLVA analysis of O-serogroups showed that the O128 strains were found in CC1 and O55 strains were grouped in CC3, the remaining CCs were contained of other O serogroups and untypeable strains (Figure 1).
4.4. OI-122 Genes
The individual OI-122 genes were detected in 93.7% of tEPEC and 42.3% of aEPEC isolates (Table 2). The distribution of individual OI-122 modules is shown in Table 3.The complete OI-122 was identified in 3 (18.75%) of tEPEC and in 1 (3.84%) of aEPEC strains. Regarding the prevalence of the modules, module 2 was the most prevalent, followed by 1 and 3. OI-122 modules were not found in 18 (42.8%) isolates.
| OI-122 Modules | Strain | ||
|---|---|---|---|
| tEPEC, n = 16 | aEPEC, n = 26 | Total, n = 42 | |
| Complete OI-122 | 3 | 1 | 4 |
| Module 1 | - | - | - |
| Module 2 | 6 | 7 | 13 |
| Module 3 | 1 | - | 1 |
| Module 1+2 | 3 | 1 | 4 |
| Module 1+3 | - | - | - |
| Module 2+3 | 2 | - | 2 |
| Absent OI-122 | 1 | 17 | 18 |
The Diversity of OI-122 Modules Among in EPEC Strains in This Study
nleB (n = 22, 52.4%) was the most frequently detected, and the prevalence of pagC, efa1/lifA, and sen genes were 19% (n = 8), 16.7% (n = 7), and 14.3%, respectively. There was a strong association between nleB and tEPEC strains (P < 0.0001). Furthermore, it was shown that efa/lifA (P = 0.008), sen (P = 0.023), and pagC (P = 0.025) had statistically significant associations with tEPEC strains (Table 2).
The EPEC strains were classified into three different groups by component analysis of the OI-122 genes in the study (Table 3). The first group included strains containing a complete OI-122 module (3 strains in tEPEC and 1 in aEPEC), an incomplete OI-122 module carrying strains classified in second group (12 strains in tEPEC and 8 in aEPEC) and the third group lack the OI-122 module (1 strains in tEPEC and 17 in aEPEC). The OI-122 genes were distributed among strains with different serogroups or even untypeable EPEC strains and there were not any clonal relationships between OI-122 genes and distinct O serogroups.
5. Discussion
The main goal of this study was to determine the prevalence and distribution of pathogenicity island OI-122 in tEPEC and aEPEC. Four principle results emerged from our data: (1) the OI-122 modules were found in all tEPEC (except one isolate), but the levels of distribution varied among the OI-122 modules observed within isolates, (2) more aEPEC (57.7%) lake of OI-122 genes and the OI-122 modules was prevalent in tEPEC than in aEPEC isolates, (3) there was statistical evidence for the existence of nleB, efa/lifA, sen and pagC genes in tEPEC genotype, and (4) there was a high degree of genetic heterogeneity among OI-122 carrying EPEC strains. These results were consistent with those previously reported; suggesting that the acquisition of complete OI-122 island, OI-122 module, and OI-122 genes are evidence of horizontal gene transfer and the evolutionary dynamics among EPEC strains (5, 15). While obtained of the OI-122 genes are associated with severe diseases and outbreaks (6), our results could not indicate the association between OI-122 module and severity of disease because all of the strains analyzed in this study were isolated from children with acute diarrhea.
A complete OI-122 gene (carrying efa1/lifA, pagC, sen, and nleB) was more prevalent in tEPEC (18.75%) than in aEPEC (3.84%). Similar results were reported by Vieira et al. and Salvador et al. (5, 15).
The efa1/lifA gene (coding for EHEC factor for adherence protein) was absent in our aEPEC strains with one exception, this gene encodes a lymphostatin, which has been shown to have adhesive potentials and inhibitory effects in lymphocyte functions (4-6). An efa1/lifA gene was found in 37.5% of tEPEC and in 3.8% of aEPEC strains in this study, the significant association with diarrhea was reported for the efa1/lifA gene (6). The efa1/lifA frequency in aEPEC strains in Japan, Brazil, Norway, Australia and New Zealand were 32.5%, 30.4%, and 28.8% respectively, which it is higher than the results in the current study for aEPEC strains (5-7).
Module 2 was the prevalent module among tEPEC and aEPEC strains in this study, which it is consistent with previous reports (5-7). This module encodes nleB (virulence factor) and sen (a putative enterotoxin) genes (16, 17). The nleB detected in the majority (87.5%) of the tEPEC strains examined, suggesting that it represents a stable acquisition of the positive clonal lineages.
The pagC gene (immunogen and bacterial survival factor within macrophages) from module 1 was present in strains carrying a complete OI-122 with all three modules and strains carrying the combination of this module and module 2. This module plays an important role (immunogen and bacterial survival factor within macrophages) in O157: H7 infections, (18, 19) but its role in EPEC infection is unknown. The heterogeneous distribution of OI-122 modules was observed in EPEC strains in this work and that of other researchers, (5-7) probably due to the acquisition of OI-122 gene contents in a modular manner.
Most of the strains (76.19%) in this study belonged to the B1 phylogenetic group and only two isolates were the B2 phylogenetic group. This typing method classifies strains according to the presence or absence of the three genes chuA, yjaA, and tspE4C2, and the only B2 group is positive for a yjaA gene (12). The significant negative association with diarrhea for the yjaA gene reported in the study of Afset et al. (6) and suggested that the presence of the yjaA gene may be a marker for low diarrheagenic potential aEPEC strains. The yjaA gene as the negative marker for diarrhea may be one possible reason for the association of B2 phylogenetic group with extraintestinal E. coli infections (20-22).
The common serogroups were O128, O111, O55 and O44 in EPEC in this study, and 50% of tEPEC and 42.3% of aEPEC strains were untypeable with the traditional O antisera. The presence of the high number of untypeable strains in this study indicates that O serogrouping is not the valuable method in the diagnosis of EPEC infections.
The extensive heterogeneity among the tEPEC and aEPEC strains in this study was found by the MLVA analysis; differences in the MLVA type were identified between strains belonging to the same phylogenetic group or O serogroups. However, two strains shared a same (M23) MLVA type (Table 2), these strains were isolated from a sister and brother, which are belong to the B1 phylogenetic group and O128 serogroup of aEPEC. Interestingly, while the OI-122 genes were absent in one of these strains, the other strain harbored the nleB gene. MLVA typing identified seven clonal complexes (7CC) in EPEC strains, but clonal relationships were not found with OI-122 genes. The MLST results of aEPEC isolates from China also showed very heterogeneity (23). Staples et al. (24) analyzed EPEC isolates by MLST and MLVA. Their isolates were highly heterogeneous. They concluded that the human enteric EPEC population might be a complex of commensal or pathogenic strains. MLVA typing of aEPEC strains from poultry suggested that poultry strains were closer to bovine strains and were less similar to patient strains (25).
In conclusion, the results of this study provide evidence for dynamic evolution regarding the OI-122 pathogenicity island in tEPEC and aEPEC strains. The OI-122 gene contents distribution in a modular manner in this study indicating that clinical, animal and cell line studies need for an understanding of the OI-122 genes roles in pathogenicity of EPEC strains. Since no specific results on the distribution of the OI-122 pathogenicity island in EPEC strains are available in Iran, the results of this study shed light on the roles of OI-122 genes in the pathogenicity and epidemiology of EPEC strains, particularly of the aEPEC strains.