1. Background
Leptospirosis is a zoonotic disease caused by pathogenic serovars of Leptospira. There are more than 200 serovars related to pathogenic strain Leptospira interrogans (1). Clinical signs vary from asymptomatic to mild and severe, with more than 500000 severe cases being reported annually worldwide (1). Due to its resemblance to other diseases (particularly malaria and dengue fever), the diagnosis cannot be made by clinical signs and thus, laboratory diagnosis is essential (2). The microscopic agglutination test (MAT) as the reference serological test is often positive in the second week; however, this test is a laborious technique, requiring two sera, live antigens that must be reactivated weekly; it also may threaten the users (1-3). Although this test has low sensitivity and specificity during the acute phase of the disease and the second sera should be considered at the convalescent phase, it is often used for prompt diagnosis (2, 4, 5).
Prompt diagnosis is critical because clinical signs may develop to severe manifestations leading to death (1). PCR from sera samples was developed to detect Leptospira during the first week of illness, and it tends to replace serological methods in endemic zones due to its sensitivity and capacity (3). PCR can detect between 102 and 103 bacteria/mL of pure culture, whole-blood, plasma, and sera samples (6).
Real-time PCR is superior to conventional PCR methods; real time is fast and simple, with excellent sensitivity and specificity; moreover, the contamination of subsequent analyses is negligent (7).
Several qPCR assays have been described; some of them amplify particular sequences of genes, which universally present in bacteria, such as rrs (16S rRNA) (8), gyrB (9) and secY (6), or genes that are restricted to the pathogenic serovars of Leptospira, e.g., lipL32 (10, 11), ligA (12), ligB (12), and lfb1 (6), or genes found in non-pathogenic strains such as 23 S rRNA (12).
Two methods of real-time PCR with the most common use are based on SYBR green technology (10) or TaqMan probes (11). lipL32 gene is the virulence factor of pathogenic species (10), which can be detected by SYBR green technology or TaqMan method (6, 10). SYBR green has lower cost and could be an important diagnostic tool, but it has some disadvantages, including its lower specificity compared to the TaqMan method, depending on fluorescent probes (6).
In Morocco, the diagnosis of human leptospirosis in the National Institute of Hygiene in Rabat always is made by serological tests, SAT (13) or ELISA IgM (5, 14). In this study, qPCR was used for the first time to evaluate its ability for the diagnosis of human leptospirosis.
2. Objectives
The aim of this study was to evaluate qPCR as a diagnostic method for human leptospirosis at the National Institute of Hygiene, Rabat.
3. Methods
3.1. Sera from Patients Suspected with Leptospirosis
Sixty-seven single sera related to 67 patients from different regions from 2004 to 2016 were examined in this study. All the sera were transferred to the National Institute of Hygiene in Rabat, Morocco, for routine diagnosis and confirmation. The main symptoms reported were icterus, abdominal myalgia, and fever. The clinicians did not consider the epidemiological information and the sampling time of patients included in this study.
3.2. DNA Extraction
A commercial QIAamp DNA Mini Kit, QIAGEN, Germany, was used to extract DNA. The total DNA was extracted using 200 µL of patients’ sera and was eluted in a final volume of 200 µL as described previously (15, 16) and according to the manufacturer’s instructions, as follows:
Twenty microlitres of proteinase K was mixed with 200 µL of lysis buffer and 200 µL of serum sample (if the sample volume was less than 200 µL, an appropriate volume of PBS would be added). Two hundred microlitres of ethanol (96 - 100%) was added and centrifuged at 6000 g (8000 rpm) for 1 min. AW1 and AW2 buffers were added and centrifuged at 6000 g (8000 rpm) for 1 min and at full speed (20,000 g; 14,000 rpm) for 3 min, respectively. Finally, DNA was eluted with 150 µL of AE buffer and then centrifuged at 6000 g (8000 rpm) for 1 min. The eluted DNA was stored at - 20°C until use.
3.3. Primer Designs
Forward and reverse primers and the TaqMan probe were designed to amplify lipL32 sequences from pathogenic Leptospira serovars. A Genesig standard kit for leptospirosis qPCR from Genesig Company was used. GenBank species included in this kit are shown in the Appendix 1 in Supplementary File. Forward and reverse primer sequences were not provided by the company.
3.4. Real-Time PCR (qPCR)
Real-time quantitative PCR was performed using the CFX96 real-time PCR detection system (Bio-Rad, USA). The amplification mixture consisted of 10 µL of master mix, 1 µL of primer/probe mix, 4 µL of RNAse/DNAse free water, and 5 µL of template DNA in a total volume of 20 µL. For DNA amplification, the program was used according to the manufacturer's instructions (Appendix 2 in Supplementary File).
3.5. ELISA IgM and IgG
IgM and IgG antibodies were determined by a commercial leptospira IgM ELISA kit and leptospira IgG ELISA kit from Nal von Minden, Germany. All sera and controls were diluted at 1:100 and were examined according to the manufacturer’s instructions and as previously reported (5).
3.6. Slide Agglutination Test (SAT)
Leptospira antigen purchased from Bio-Rad (Marnes-la-Coquette, France) was used according to the manufacturer’s instructions and as previously described (13, 17). The antigenic suspension was homogenized and 15 µL volumes of each undiluted serum were added to the antigenic suspension on a glass slide, and the agglutination was observed under direct light for four minutes.
4. Results
4.1. ELISA IgM and SAT
Of 67 sera included, 39 (58.2%) and 48 (71.6%) were positive by ELISA IgM and SAT, respectively (Table 1). Of them, 61 sera had been diagnosed previously by SAT and ELISA IgM (5, 13, 14).
| Number of Serum Samples | No. (%) of Sera Positive by: | |
|---|---|---|
| SAT | ELISA IgM | |
| 67 | 48 (71.6) | 39 (58.2) |
4.2. ELISA IgG
Of 17 sera subjected to ELISA IgG, only one serum had a positive result (Table 2). The positive serum by ELISA IgG was also positive by ELISA IgM and SAT while it was negative by qPCR.
| Number of serum samples | No. (%) of Sera Positive by: | |||
|---|---|---|---|---|
| SAT | ELISA IgM | ELISA IgG | Real-Time PCR | |
| 17 | 15 (88.24) | 10 (58.82) | 1 (5.88) | 3 (17.64) |
4.3. qPCR
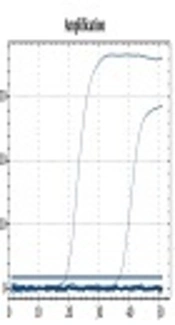
Thirty-six sera were subjected to qPCR, and only three sera had positive results (Table 3 and Figure 1). The positivity of PCR was 8.33% compared to SAT with 41.66% and ELISA with 27.77%. Of 36 sera tested by qPCR, 19 had negative results by ELISA IgM and SAT (5, 14). All negative sera by ELISA IgM and SAT were also negative by qPCR, and the three positive sera by PCR were positive by ELISA IgM and SAT, as well.
| Number of Serum Samples | No. (%) of Sera Positive by: | ||
|---|---|---|---|
| SAT | ELISA IgM | PCR Real Time | |
| 36 | 15 (41.66) | 10 (27.77) | 3 (8.33) |
4.4. Geographical Distribution of Cases
Sera were obtained from nine regions from 2004 to 2016 (Figure 2). High incidences were observed in Sidi Qacem, followed by Meknes (Table 4). During this period, sera related to males were 46 (68.68%) compared to 21 (31.34%) related to females.
| Region | No. (%) |
|---|---|
| Beni Melal | 1 (2) |
| Meknes | 21 (31) |
| Agadir | 1 (2) |
| Rabat | 2 (3) |
| Taza | 2 (3) |
| Sala | 2 (3) |
| Tanger | 2 (3) |
| El Jadidah | 9 (13) |
| Sidi Qacem | 27 (40) |
5. Discussion
Since 2004, ELISA IgM and SAT have been routinely used in our laboratory for the diagnosis of human leptospirosis (5). Although the sensitivity of serological tests increases in the second week of the illness (2), serological methods are still being used due to their facility, sensitivity, and availability (4). SAT and ELISA IgM seem to be superior at the acute phase to MAT as the reference serological method (2, 4), and they are suggested for the diagnosis of leptospirosis (2, 18). PCR has more sensitivity in the first days of illness due to its ability to detect 2 - 20 genomics of leptospires from sera and 10 genomics from urine (11, 16). However, low positivity during the course of the disease was observed (11, 18-20).
In Morocco, MAT is suggested for the diagnosis (21); however, the test is not available in Morocco and it is conducted in the Pasteur Institute of Paris (21). Real-time PCR was reported in animal leptospirosis in one study (22). The laboratory of Bacteriology at the National Institute of Hygiene in Rabat is the first laboratory in Morocco that has the ability to conduct real-time PCR for human leptospirosis.
The positivity of SAT and ELISA IgM for all 67 sera was 48 (71.6%) and 39 (58.2%), respectively. In the case of the unavailability of the reference serology test (MAT), ELISA IgM and SAT can detect leptospirosis at the acute and convalescent stages, with high sensitivity as reported (2, 4, 19, 23, 24). Therefore, we suggest that patients who had negative results by ELISA IgM and SAT might have not leptospirosis.
De Abreu Fonseca et al. indicated that SAT and ELISA IgM had adequate sensitivity at the acute phase and that SAT could detect leptospira antibodies earlier that ELISA IgM (18). Brandao et al. reported that the positivity of SAT, ELISA IgM, and MAT at the acute phase was 57%, 53%, and 34%, respectively; however, the positivity reached 99% in all tests from the 15th day of the illness (2).
ELISA IgG had the lowest positivity (5.88%) among others including SAT (88.24%), ELISA IgM (58.82%), and real-time PCR (8.33%). The low positivity of ELISA IgG might be due to that sampling was done before the development of IgG antibodies. Cumberland et al. reported that IgG antibodies appeared after IgM, and maximum titers of IgG can be detected at the convalescent phase of the illness (25).
In our study, of 36 sera subjected to qPCR, only three sera had positive results. A lower positivity was observed in qPCR compared to ELISA IgM and SAT. Although the date of sampling was not reported by clinicians, sera that were negative by qPCR and positive by ELISA IgM and SAT might be due to the late sampling conducted after leptospires were cleared from the blood. Perwez et al. reported that PCR was superior in the first week of illness to ELISA IgM, but a low positivity was observed from the 8th day, and during 13 - 15 days, ELISA IgM was superior to PCR (24).
De Abreu Fonseca et al. compared PCR with ELISA IgM, and reported that ELISA IgM had more sensitivity; however, PCR was most sensitive in initial sera samples presenting no specific antibodies detectable by any of the serological methods tested (18). Therefore, it was suggested that the positivity of the diagnosis increased when PCR was used with serological tests (18, 23).
In addition to late sampling, several reasons may lead to low positivity in PCR, such as substances (e.g. urea, creatinine, and hemoglobin derivates) that may inhibit DNA amplification with leptospiral primers (18). Moreover, other possible reasons include the absence of the organisms in the blood (26), the microbial counts of about five cells that were too low to be detected (20, 26), and degradation of DNA due to prolonged sample storage and several sample thawing (26, 27).
Moreover, using the serum for the diagnosis may lead to less positivity. Kositanont et al. observed that the positive rate for DNA detection was higher in buffy coat (peripheral white blood cells) than in plasma and serum (28). Stoddard et al. also indicated the same findings (11). For this reason, serum seems to be a non-optimal specimen for real-time PCR (11); however, serum gave high positivity than whole-blood (20, 27).
Our findings revealed that males had higher rates of leptospirosis than females, which may be due to the occupation factor (1, 19). The majority of cases were observed in Sidi Qacem region, followed by Meknes. The high incidence in Sidi Qacem was indicated earlier (5, 14), and high incidences in the Meknes region was related to an outbreak occurred in 2004 as reported previously (5, 29).
Due to the limitations of resources, we could not perform blood cultures on patient’s samples and MAT; therefore, we were unable to assess the sensitivity of the tests used in this study. Other limitations should be recognized such as unavailability of the time of sampling, second sample , and infecting serovars . We recommend conducting the diagnosis from buffy coat and urine specimens. Urine is a useful sample for testing of leptospirosis because the bacteria are present in the blood only in about the first week after the onset of symptoms, but they can be detected in the urine for several weeks (1). Moreover, multiplex PCR (mPCR) should be considered when it is available; the use of two sets of primers in mPCR can increase the sensitivity and specificity of the test (30).
5.1. Conclusions
Our study agrees with other studies that PCR is not useful for the diagnosis during the course of leptospirosis, and ELISA IgG cannot be used for early diagnosis. ELISA IgM and SAT are useful and rapid and do not need highly experienced labor; moreover, they are available in low-income countries and in less-equipped laboratories, and seem to be useful for the diagnosis of human leptospirosis.