1. Background
HIV-related death rates have been reduced significantly due to highly active anti-retroviral therapy regimen (HAART) (1). However, the efficiency of antiretroviral therapy (ART) is often limited due to the emergence of drug resistant HIV strains (2, 3). HIV-1 resistance to antiretroviral drugs is a common problem that limits the effectiveness of ART (4). Drug selection pressure (acquired resistance) or person-to-person transmission (transmission resistance) are two pathways leading to drug resistant HIV-1 patient (5). In developing countries, the main reasons for the occurrence of drug resistance are financial constraints; a lack of testing to detect HIV drug resistance mutations (DRMs) before treatment; an interruption of ART because of disruptions in the supply of anti-HIV drugs; and the inappropriate administration of antiretroviral regimens (6). Currently, it is highly recommended that a drug resistance test be used to detect resistance to anti retrovirals when monitoring HIV treatment and to make the appropriate changes in the medication regimen (7). The current methods for testing HIV drug resistance include phenotypic drug susceptibility tests for measuring in vitro HIV-1 drug inhibition and genetic tests for identifying the mutations that lead to drug resistance (8).
The anti-HIV-1 drugs are available in six approved classes, including nucleoside and nucleotide reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), fusion inhibitor, CC chemokine receptor 5 antagonist and integrase inhibitor (5). The protease, enzyme code 3.4.23.16, is a homodimer aspartyl enzyme whose subunits have 99 residues separately. During viral maturation in HIV-1 replication cycle, the protease enzyme is responsible for breaking selectively and proficiently the peptide bonds in the viral polyproteins of gag and pol genes (9). Data on the structure and function of the HIV protease enzyme and the experimental settings to check the actions of other aspartyl protease inhibitors have been fundamental in designing new inhibitors for HIV-1 protease enzyme (10). As has been mentioned extensively in clinical trials and cohort studies (11), the use of PIs in combination with retroviral drugs leads to an increased suppression of the HIV-1 virus (12, 13). Therefore, HIV-1 protease (PR) is a key target in management of HIV-1 infected patients. As one of the most important drug targets, the PR is exposed to strong selective pressure during ART treatment. The resistance to protease occurs by the accumulation of certain major or minor DRMs (14). Mutations are classified into major mutations, which develop extreme resistance to PIs, and minor mutations, which cause low-level resistance to PIs (15).
2. Objectives
The present study was conducted to obtain the complete sequence of the protease gene and to detect the associated DRMs in treated and untreated Iranian patients infected with HIV.
3. Methods
3.1. Study Population
This cross-sectional study was conducted on the blood samples of 50 HIV-1 positive patients sampling was performed by Mashhad and Tehran health centers. Patients were divided into 2 groups. The first group consisted of 25 HIV patients who did not receive any treatment. The second group included 25 HIV patients who had received a PI regimen, specifically Kaletra (Lopinavir and Ritonavir), Atazanavir, Ritonavir, or Darunavir for at least 1 year; the CD4 count had been determined by flow cytometry.
3.2. Measurement of Laboratory Parameters
Peripheral blood mononuclear cells (PBMCs) were collected from blood samples using the Ficoll-Paque method. Proviral DNA was extracted using the Genet Bio kit (Seoul, South Korea) according to the manufacturer’s instructions. A nested polymerase chain reaction (Nested PCR) was performed in two rounds. In the first round, 2.5 μL of 10X PCR buffer, 2 μL of 25 mmol/L MgCl2, 0.5 μL of 2.5 mmol/L dNTP, 1 mmol/L of 10 pmol/μL outer primers, 0.3 μg of Taq DNA polymerase (Genet Bio, Seoul, South Korea), and 3 μL of extracted DNA provirus were mixed together, and distilled water was added to a final volume of 25 μL. Tubes were placed in a thermocycler machine (Applied Biosystems, Foster City, CA, USA) with the following cycling program: denaturation at 95°C for 3 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 40 s, extension at 72°C for 35 s, and the final extension of strands at 72°C for 3 min. The second amplification using inner primers was similar to the first one except that only 1 μL of PCR product was used and the following cycling program was used: denaturation at 94°C for 4 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 40 s, extension at 72°C for 40 s, and 3 min final extension step of at 72°C. We used previously published primers to amplify PR gene (16). The sequence of the primers used in this study is presented in Table 1. The length of the final product was 424 nucleotides. Paired-end sequencing was performed using the genetic analyzer ABI 3130xl (Applied Biosystems, USA).
| Name | Primer Direction | Nucleotide Sequence (5’ to 3’) |
|---|---|---|
| DRPRO5 | Forward (external) | AGA CAG GYT AAT TTT TTA GGG A |
| DRPRO2L | Reverse (external) | TAT GGA TTT TCA GGC CCA ATT TTT GA |
| DRPRO1M | Forward (internal) | AGA GCC AAC AGC CCC ACC AG |
| DRPRO6 | Reverse (internal) | ACT TTT GGG CCA TCC ATT CC |
3.3. Drug Resistance Analysis
Chromas 2.6.2 software was used to read sequenced nucleotides. The sequences were analyzed and interpreted by the Stanford University HIV Drug Resistance Database (HIVdb) [http://hivdb.stanford.edu] (17) to determine DRMs and viral subtypes.
3.4. GenBank Accession Numbers of PR Sequences
The nucleotide sequences of the PR sequences of HIV received from this study (424 bp) were submitted to the Gen Bank database and are accessible under the accession numbers MG971405 to MG971420.
3.5. Ethical Considerations
The research was performed totally in adhesion with the principles of the Helsinki declaration on human research. Prior to the study, the Medical Ethics Committee of Mashhad University of Medical Sciences reviewed and approved study protocol (IR.MUMS.fm.REC.1395.105). The study objectives were presented to all participating patients, who were then complete a written consent form.
3.6. Statistical Analysis
Descriptive analysis (frequency, percentage, mean, standard deviation) were performed with Statistical Package for Social Science version 11.5.
4. Results
Patients who participated in this study aged ranging from 11 - 57 years, (mean age of 40.1 years ± 10.5 years, median 41 years). 33 out of the 50 participants, (66%) were male and 17 (34%) were female. The mean count of CD4 cells in the samples was 335.6 ± 304.3 cells/mm3 ranging from 50 - 1324 cells/mm3 (median of 284 cells/mm3).Based on the data recorded in the patient files, 52% of the HIV patients were infected by drug abuse), 32% by sexual contact, 4% by infected blood products, 6% by mother-to-child transmission, and 6% by an undisclosed mechanism (Table 2).
| PI-Naïve (n = 25) | PI-Experienced (n = 25) | Total (n = 50) | |
|---|---|---|---|
| Age (y) | 41.04 ± 10.63 | 39.20 ± 10.53 | 40.1 ± 10.51 |
| CD4 cells | 279.3 ± 317.4 | 395.3 ± 280.0 | 335.68 ± 304.391 |
| Gender | |||
| Male | 16 (64) | 17 (68) | 33 (66) |
| Female | 9 (36) | 8 (32) | 17 (34) |
| Reported transmission route | |||
| Intravenous drug use | 11 (44) | 15 (60) | 26 (52) |
| Sexual contact | 10 (40) | 6 (24) | 16 (32) |
| Infected blood products | 2 (8) | 0 | 2 (4) |
| Mother-to-child transmission | 1 (4) | 2 (8) | 3 (6) |
| An undisclosed mechanism | 1 (4) | 2 (8) | 3 (6) |
aData represented as mean ± SD or frequency (percentage).
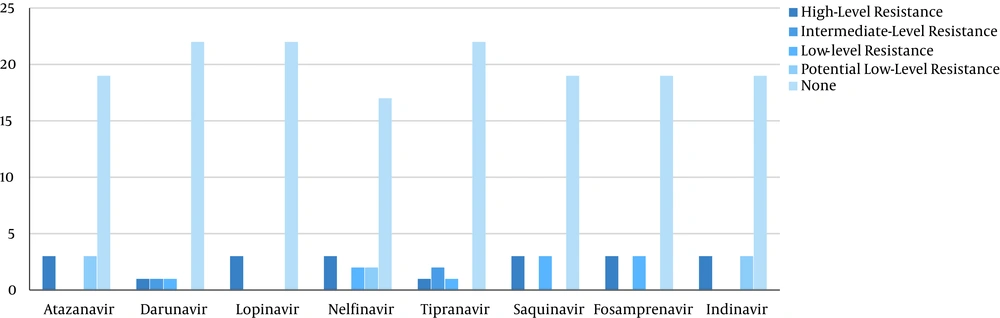
In the group that had received treatment, 68% (17) of the participants did not have any resistance mutations and, thus, were sensitive to PIs. On the other hand, 32% (8) had at least one resistance mutations to PI drugs. Three individuals had high-level resistance, 2 had moderate-level resistance, 6 had low-level resistance and 4 had potential level resistance. This adds up to 15 because some individuals had multiple resistance level to various drugs. All participants who were not under treatment were susceptible to PIs.
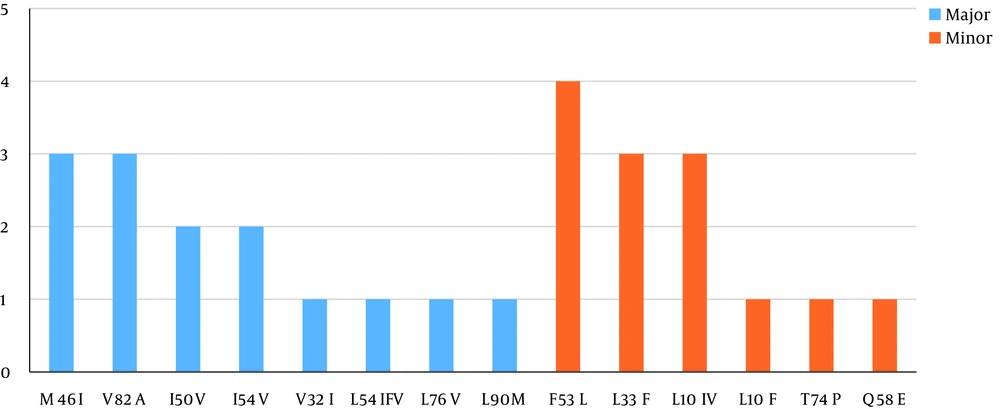
The highest resistance was related to nelfinavir (28%) and the lowest resistance was to both darunavir and lopinavir (12%).The significant major mutations observed in the treated patients that were found to induce drug resistance were M46I and V82A (12%), I50V and I54V (8%), V32I, L54IFV, L76V, and L90M (4%). The minor mutations in these patients that caused resistance were F53L (16%), L33F and L10IV (12%), and Q58E, T74P, and L10F (4%) (Figure 1). The subtype analysis that was performed simultaneously with the analysis of the mutations showed that 94% of the samples were of subtype CRF35_AD, and 6% were of subtype A. The frequency of the resistance levels to PIs is shown in (Figure 2).
5. Discussion
To prevent the transmission of drug-resistant HIV strains, the rapid detection of resistant strains in among patients and the development of strategies to cope with resistant strains circulating in the community are important issues that should be addressed in the public health system. In this study, samples from patients infected with HIV who did not receive any ART were tested for the presence or absence of protease gene mutations associated with drug resistance; the results showed no resistance mutations among these patients. Whereas in the samples from patients infected with HIV who had received PIs, 32% had drug resistance mutations to PI drugs that are available in Iran, including Kaletra (lopinavir and ritonavir), Atazanavir, Ritonavir, and Darunavir. Baesi and Hamkar in Tehran, Iran found that 40% and 45% of samples from HIV patients had resistance mutations to PIs, respectively (18, 19); these results were consistent with the percentage of resistance mutations seen in the present study. However, Naziri in Gorgan, Iran reported that 5% of the samples had PI resistance mutations (20). In this study, the most common PI resistance mutations were at codons 82 and 46 (23%), while the most common PI mutations reported by Baesi and Hamkar were at codon 90 (18, 19). Globally, studies have reported the prevalence of PI-resistant HIV as follows: North America up to 12.9%, Europe (10.9%), Latin America (6.3%), Africa (4.7%) and (4.2%) in Asia (21).
To some extent, all PIs have cross-resistance (22). This means that the resistance mutations that occur in the presence of one PI, reduce the effectiveness of other PIs. There is evidence that HIV strain that has reduced susceptibility to Kaletra (lopinavir and ritonavir) shows a high-level resistance to Indinavir (Crixivan) and Ritonavir, a moderate-level resistance to Amprenavir (Agenerase), while is susceptible to Saquinavir (Invirase) (22). The PI treatment regimen containing Kaletra (lopinavir/ritonavir) is widely used. The World Health Organization (WHO) has recommended the Kaletra regimen to be used as the first-line combination ART for children under three years old infected with HIV in the developing world; however in many cases the Kaletra regime is used as second-line ART (23). To date, decreased susceptibility to Kaletra has been mainly associated with mutations in 46I, 54V, 71V, 82A, and 84V codons (24-26). The Atazanavir resistance can occur due to numerous mutations with different effects; among these I50L, I84V, and N88S cause high level resistance. When ritonavir is used to boost atazanavir, increased number of mutations are required to cause impaired antiviral effect (27). Phenotypic studies on several PI resistance mutations showed that V32I, I50V, I54M, L76V, and V82F largely decreases the susceptibility to Darunavir (28). Ritonavir is not currently used for its own antiviral effect, and instead is mainly used to boost the blood levels of other PIs (29). High-dose ritonavir monotherapy, which was used in the past, caused mutations similar to those associated with Indinavir (30). Our study showed that the mutations associated with Kaletra occurred most frequently at codons 46, 54 and 82. The mutations associated with Atazanavir occurred at codons 46 and 76 and for Darunavir, mutations occurred at codons 32, 50 and 76.
Our findings showed that a vast majority (94%) of studied patients had HIV-1 CRF35_AD subtype. This finding is consistent with previous reports, which recognized the recombinant subtype CRF35-AD as the most prevalent variant in Iran (6, 31-36). The prevalence of CRF35_AD among HIV patients who were infected by syringe (injecting drugs) has been observed across Iran (6).
Some limitations of the study should be noted. First like many other patients-based studies some lacking data might exist due to the incomplete patients’ files. Second we checked genotypically resistant mutations, it would be helpful if we could add phenotypical assays however, we think the present study provides remarkable data on resistance mutations regardless of the applied method and finally larger sample size could definitely improve the study.
5.1. Conclusion
Remarkable treatment outcomes and long-term effectiveness of the antiretroviral drugs among HIV-1 patients could be achieved by analyzing HIV-1-drug resistance profile before PI treatment onset. High prevalence of drug resistance mutations was reported in our investigation in our HIV patients. According to the present results, it can be concluded that the proper direction of therapeutic plans and restrictions of transmission of resistant variants should be considered as strategies to control and prevent emerging resistant strains.