1. Context
The incidence of invasive fungal infections (IFI) caused by unusual pathogens is on a growing trend, partly driven by the increased population of immunocompromised patients. Among other Candida species, widespread resistance to conventional antifungal drugs, high treatment failure, and high mortality rate are the characteristics of fungemia due to C. auris. (1). The word ‘auris’ is derived from Latin and means ‘ear’. Candida auris is an emerging fungal superbug characterized by high multidrug resistance and as such is regarded as a serious threat and source of concern among healthcare givers.
During 2004 - 2006, unconventional yeast isolates phenotypically just like Candida haemulonii were reclaimed in culture from scientific samples collected from 23 patients in five Korean hospitals. Almost all of these isolates were significantly less susceptible to amphotericin B (AMB) or even fluconazole (FZU) compared to most other Candida species (2). In 2009, yeast isolates from the external auditory canal of Japanese patients were identified to be Candida auris using sequence analysis of the nuclear rRNA gene (3). In the same year, about 15 C. auris isolates were retrieved from otitis media patients in South Korea (4). In 2011, it was described that three bloodstream infections (BSIs) were caused by C. auris in South Korea, and C. auris was later isolated from patients in India, South Africa, Kuwait, Venezuela and Pakistan (5-7). These isolates have been reported in Germany and Norway as well (8).
The first episode of candidemia outbreak due to Candida auris in Venezuella occurred between early 2012 and mid 2013 in pediatric intensive care unit (ICU) (9). In Columbia, a few episodes of the infection have been reported sporadically since year 2013. Between 2015 and 2016, less than 30 isolates of the yeast were verified in Barranquilla city of Columbia. Similarly, in Cartagena, another episode of outbreak occurred in pediatric ICU patients in August 2016. It involved disseminated yeast infections caused by initially misidentified yeast species that were later verified to be C. auris.
Candida auris infections have also been reported in several parts of England since 2013. Between 2015 and 2016, a cardiothoracic center experienced the episodes of outbreak (9, 10). The isolated species were found to be evolutionary and related to some rare Candida species such as C. haemulonii. Candida haemulonii or Torulopsis haemulonii is recognized as one of the rare yeast species that can be isolated from human clinical samples (11). The draft genome of C. auris has a genome size of approximately12.3 Mb (12, 13). The phospholipase (PL) activity and secreted proteinase were detected in 37.5% and 64% of the tested isolates, respectively (14, 15).
Comparison of whole-genome sequencing (WGS) data demonstrated that C. auris is phylogenetically closely related to C. lusitania, a species recognized for innate anti-fungal resistance. Using WGS, three different amino have been identified in the ERG11 gene of C. auris. These substitutions were exclusively associated with geographic clades, namely Y132F from Venezuela, F126T from South Africa and Y132F or K143F from India and Pakistan. Each mutation was linked to isolates from a different country, indicating that resistance to FCZ might be acquired rather than inherent (6, 9, 16).
The emergence of Candida auris as a significant pathogen in tertiary and pediatric centers is alarming. Recently, about 8.6% to 30% of candidemia cases have been reported in India (17). Healthcare-based epidemics caused by C. auris have occurred across Asia and South America, as highlighted in a 2016 clinical warning (18). Hospital-acquired infections are gaining momentum with estimates of about 99000 deaths a year, according to the Centre for Disease Control (CDC) in the USA (19). Hospital-acquired IFI is of public health concern and candidemia is becoming very relevant in European countries (13). In India, the national prevalence of candidemia in ICUs due to C. auris was around 5% and as high as 30% of the Candida isolates were identified to be C. auris in some centers (20, 21).
2. Who Is at Risk?
Available data show that the risk factors for acquiring C. auris infections are not very different from those for other Candida species. These include immunosuppressive conditions such as diabetes mellitus (DM), cancers and chemotherapy, the presence of central line catheters, the use of broad-spectrum antibiotics, neutropenia, total parenteral nutrition (TPN), hemodialysis, blood transfusion, major surgery within one month, critical care, previous therapy with antifungal agents within one month, concomitant bacteremia or candidemia, candiduria, indwelling urinary catheter and chronic kidney diseases (9).
C. auris infection is defined as any clinical case with clinical signs and symptoms of infection and a positive culture sample from a non-sterile site (e.g., urine or sternal wound,) requiring treatment with antifungal agents (10). The emerging MDR yeast pathogen Candida auris has attracted considerable attention as a source of healthcare-associated infections (9). This pathogen has been associated with life-threatening invasive diseases such as candidemia and wound infections. About 20% of cases of colonization with C. auris resulted in candidemia among ICU-hospitalized patients in the UK (7, 22, 23). Other reports showed that mortality following C. auris candidemia ranged from 30% to 60%, involving larger groups of adult patients (24).
Transmission of infection was observed in intensive care, surgical, medical, neonatal, oncologic, and pediatric wards, which were mutually exclusive with respect to healthcare staff. Biofilm formation is an important phenomenon for C. albicans pathogenicity and is associated with patient death. Genome analysis is equally important in identifying the key proteins involved in slime formation (22, 25, 26). The clinical resistance of C. auris to FCZ is distinctly alarming. Alterations at azole-resistance codons have been detected in both C. albicans and C. auris isolates by comparing ERG11 amino acid sequences between the two species (6).
3. Mode of Transmission Candida auris
• Person to person
• Spread through contact with contaminated environmental surfaces
• Direct contact with contaminated fomites (such as blood pressure cuffs, stethoscopes, and other equipment in contact with the patient)
• Indirect transmission from the hands of health care staff
• Invasive procedures or exposure to an indwelling device
4. Candida auris Virulence Factors
• Histidine kinase-2 component system
• Iron acquisition
• Tissue invasion
• Enzyme secretion
• Phospholipase and proteinase production
• Multidrug efflux
• Genes/pathways involved in cell wall modeling
• Nutrition acquisition
• Salt tolerance and cell aggregation
• Thermotolerance up to 42°C
• Biofilm formation (15)
5. Clinical Conditions and Mortality
C. auris has been identified as an agent of serious invasive fungal infections associated with high mortality rate (up to 72%) (6, 18, 23).
It has been isolated from various clinical conditions including:
• Candidemia
• Urinary tract infection (UTI)
• Otitis
• Surgical site infections
• Skin lesions associated with catheter insertion
• Cardiac muscles infection
• Central nervous system infections such as meningitis
• Bone infections (BI)
• Burns wound infection and colonization (9).
6. Diagnosis of a Candida auris Infection
• Fungal culture of clinical specimen the affected site such as blood, pus and body fluids,
• Identification of the yeast isolates based on the phenotypic characteristic such as oval yeast cells without pseudohyphae or germ tube formation.
• C. auris does not form chlamydospore
• The pink colony on CHROMagar Candida medium
• Growth at 37°C and 42°C
• Assimilated N-acetylglucosamine (NAG) (C. auris isolates from India)
• Ability to ferment glucose, weak fermentation of sucrose and trehalose, but with inability to ferment galactose, maltose, lactose, or raffinose
• Starch formation, urease activity and diazonium blue B reaction are negative
• Commercially available biochemical-based tests
• VITEK-2 (misidentified isolates as C. haemulonii and C. famata)
• API20C (misidentified them as R. glutinis and C. sake)
• MicroScan AutoSCAN 4 and MicroScan Walkaway
• Molecular identification (ITS and D1/D2 regions)
• The phenotypic divergence of these- isolates (M13 and AFLP typing)
• Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) (2, 9, 27).
| Diagnostic System | Comment |
|---|---|
| API-20C glutinis, Candida | May misidentify Candida auris as Rhodotorula sake, or Saccharomyces cerevisiae |
| Vitek-2 Candida famata | May misidentify C. auris as Candida haemulonii or |
| MALDI-TOF the database | Will identify C. auris if appropriate sequences are in |
| DNA sequencing subunit rRNA | Sequencing of the ITS and D1-D2 domain of the large gene has been performed most commonly |
| Clinical and laboratories standards Institute broth microdilution method | May give falsely elevated caspofungin MICs |
| Etest | May give most consistent results |
a Abbreviation: MIC, minimum inhibitory concentrations.
| Isolate No. | Species Tested | API 20C AUXa | BD Phoenixb | Vitek-2c | MicroScand |
|---|---|---|---|---|---|
| 1 | C. auris | R. glutinis | C. catenulate | C. haemulonii | C. famata |
| 2 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. famata |
| 3 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. famata |
| 4 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. lusitaniae |
| 5 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. guilliermondii |
| 6 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. famata |
| 7 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. guilliermondii |
| 8 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. parapsilosis |
| 9 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. guilliermondii |
| 10 | C. auris | R. glutinis | C. haemulonii | C. haemulonii | C. guilliermondii |
| 11 | C. duobushaemulonii | R. glutinis | C. parapsilosis | C. haemulonii | C. guilliermondii |
| 12 | C. duobushaemulonii | R. glutinis | C. parapsilosis | C. haemulonii | C. guilliermondii |
| 13 | C. haemulonii | R. glutinis | C. haemulonii | C. haemulonii/K. ohmeri | C. catenulata |
| 14 | C. duobushaemulonii | R. glutinis | C. parapsilosis | C. haemulonii | C. parapsilosis |
| 15 | C. haemulonii | R. glutinis | None | C. haemulonii/K. ohmeri | C. parapsilosis |
a Identification at 48 and 72 hours of incubation; API 20C AUX does not have C. auris, C. haemulonii, or C. duobushaemulonii in its library.
bC. haemulonii is in the BD Phoenix library, but C. auris and C. duobushaemulonii are not.
c The Vitek-2 library has C. haemulonii but not C. auris or C. duobushaemulonii.
d MicroScan does not have C. auris, C. haemulonii, or C. duobushaemulonii in its library (28).
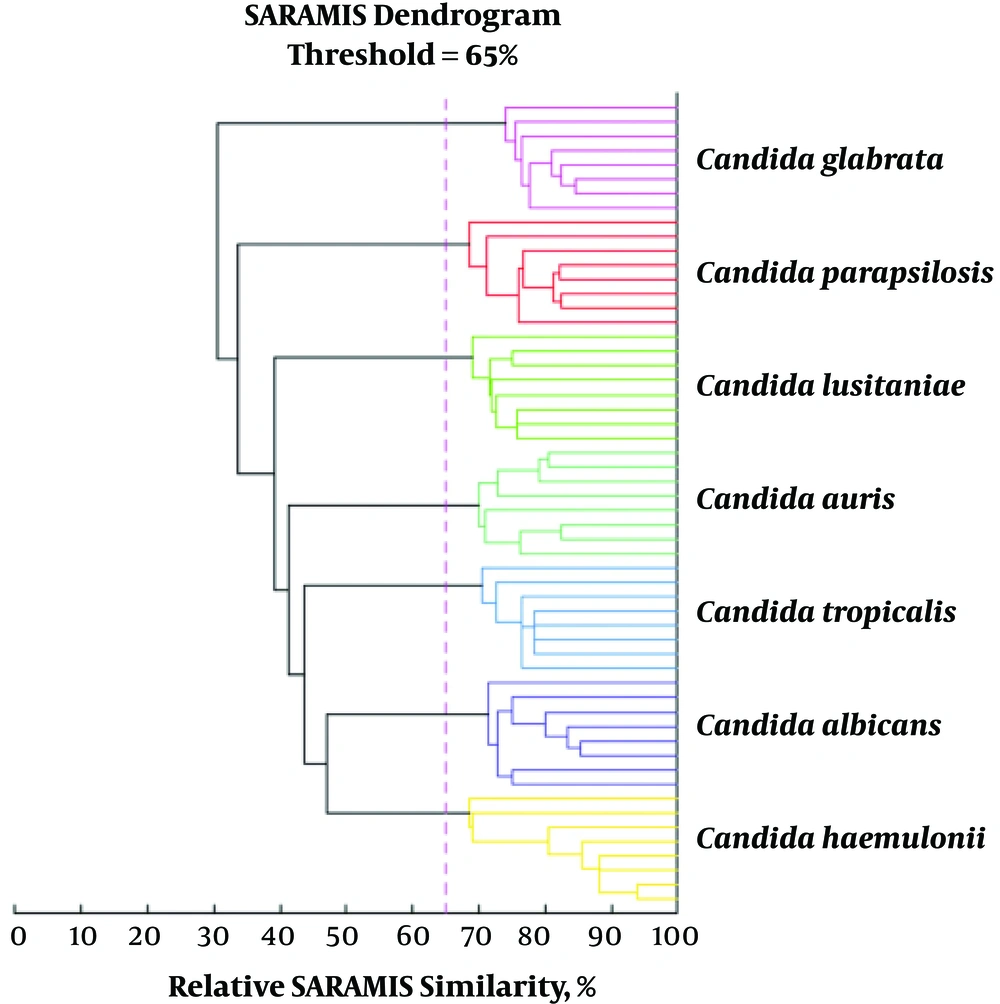
Cluster based on the relative similarity of spectra from representative isolates for the most common yeast species present in the VITEK MS database showing clear distinction between Candida species. A similarity below 65% for two spectra means that the strains belong to different species (29)
7. Decolonization
To date, clinical studies have shown that colonization with C. auris is tenacious and difficult to eradicate making infection prevention and control strategies even more important. For patient decolonization, the following methods have been suggested:
• Twice daily 2% chlorhexidine gluconate wipes twice daily or 4% chlorhexidine aqueous formulations on skin.
• Oral decolonization can be achieved by using 0.2% chlorhexidine mouthwash or 1% chlorhexidine dental gel in patients on ventilator support.
• Use of oral nystatin in oropharyngeal colonization.
• Chlorhexidine-impregnated protective disks for central vascular catheter exit sites (9).
8. Drug Resistance
The commonly used methods for testing antifungal sensitivity are the CLSI-BMD, E-test method and the VITEK-2 antifungal susceptibility test (30-32). There are reports of C. auris resistance to the major classes of anti-fungal drugs (i.e., azoles, polyenes, and echinocandins). For the purpose of epidemiological studies, the CDC has maintained the conservative breakpoints already in use for other Candida spp. than C. auris. The breakpoint sets were ≥ 32 µg/mL for FCZ, ≥ 2 µg/mL for AMB, ≥ 2 µg/mL for voriconazole (VOR), ≥ 128 µg/mL for flucytocine (FLU) and ≥ 8 µg/mL for the echinocandins. Resistance to FCZ exceeds 90%, while resistance to VOR may approach 50% among isolates of C. auris. Low MICs for the newer triazoles, such as posaconazole and isavuconazole, suggest that these antifungals may be effective against C. auris.
Resistance to AMB has been reported in up to 35% of isolates, while resistance to echinocandins has been between 2% - 8%. About 4% of isolates have been resistant to all the three major classes of anti-fungal drugs (20). SCY-078 is a triterpene glucan synthase inhibitor (GSI) that has exhibited both in-vitro and in-vivo activity against the most common Candida species (antifungal/antibiofilm), including echinocandin-resistant isolates. This drug is the only 1, 3-glucan synthase inhibitor with both oral and intravenous (IV) formulations in development.
It can be concluded that SCY-078 is a promising novel antifungal agent against Candida auris, although further investigations are warranted (15, 33). Combination therapy coupled with VOR and micafungin highlighted synergistic activities towards multidrug-resistant C. auris, indicating a great substitute approach to handle its antifungal drug resistance (34).
9. Case Reports
1- The first report of donor-derived C. auris transmission was in a lung transplant recipient. A 71-year-old male with end-stage chronic obstructive lung disease due to idiopathic pulmonary fibrosis (35).
2- Fungal otomastoiditis is a rare disease, which can be fatal in immunocompromised patients. Recently, there have been increasing cases of otologic infection caused by Candida auris (36).
3- A case report of vulvovaginal candidiasis caused by C. auris characterized by its virulence traits and drug resistance profile using molecular methods (37).
4- A rare case of fungal pericardial effusion caused by Candida auris in a patient with chronic liver disease (CLD) (38).
10. Conclusions
Numerous difficulties have been reported with regards to the identification of C. auris. Identification based on molecular methods is not commonly performed in diagnostic laboratories, leading to the underestimation of the actual incidence of C. auris infections. Currently, the diagnosis of C. auris infections should be confirmed using acceptable methods such as MALDI-TOF or molecular identification techniques like polymerase chain reaction (PCR), sequencing and amplified AFLP-PCR (39). The inherent sturdiness of C. auris to survive and persist in the environment and its high transmissibility make its infections highly tenacious. To date, clinical studies have revealed that colonization is difficult to eradicate and tends to persist, making infection prevention and control strategies even more important. Poor susceptibility to chlorhexidine could be a possibility, but not evident enough to establish whether C. auris is susceptible or resistant to chlorhexidine, which necessitates further studies in this area. However, methods such as ultraviolet light (UV), hydrogen peroxide vaporization and halide-based agents are shown to be effective for environmental cleaning.
Attention should be given specifically to the cleaning of non-disposable devices (e.g., blood pressure cuffs, thermometers and ultrasound machines), beds and surrounding areas occupied by an infected/colonized patient. Finally, it is pertinent to emphasize that C. auris is emerging as an important nosocomial pathogen in many centers in the world and this obliges developing rapid and reproducible methods for its identification and typing. Therefore, further evaluation of MALDI-TOF MS as an acceptable typing method for this yeast is warranted. Effective methods of decolonization, surface decontamination and hand hygiene are essential in preventing nosocomial infections, especially outbreaks in healthcare settings.