1. Background
An increase in the global incidence of drug-resistant Mycobacterium tuberculosis (MTB) infection threatens appropriate TB prevention, diagnosis, treatment, and case management. Therefore, there is a critical need in the healthcare system for efficient approaches to rapidly identify drug-resistant cases of MTB (1). According to the world health organization (WHO), multi-drug-resistant tuberculosis (MDR-TB) is the strain that does not respond to at least isoniazid and rifampin, which are among the most powerful first-line anti-TB drugs (2). However, taking multiple drugs concomitantly can successfully treat patients and limit the growth of MDR strains (3). As it is well-known that random genetic mutations in specific genes encoding either the target of the drug, are involved in drug activation and confers resistance to MTB isolates (4, 5). Several mechanisms of TB drug resistance have been well characterized. Point mutations, deletions, or insertions have been described for all of the first-line drugs (isoniazid, rifampin, pyrazinamide, ethambutol, and streptomycin), and for the several second-line and newer drugs (5). Thus, the establishment of molecular assays allows the rapid detection of drug resistance in clinical MTB isolates (6).
Isoniazid (INH) is a prodrug and a catalase-peroxidase encoded by katG gene, associated with peroxidase activity, activates it to be converted to a toxic substance that affects mycolic acid biosynthesis (7). Between 40% and 95% of INH resistant clinical MTB isolates have mutations in katG gene, 75% to 90% of which are located in codon315, with 10% to 25% of mutations located in other katG loci (8). Mutations in inhA (less than 10%) and ahpC gene are rare (9). Accordingly, because of several resistance-associated genes and regions, investigation of INH resistance is not quite straightforward. The S315T mutations in the katG gene occur more frequently in MDR strains and are associated with high-level resistance (10). Rifampin (RIF) resistance is considered a primary/surrogate indicator for MDR-TB and XDR-TB, since mono-resistance to INH is common; however, MDR strains refer to the strains that contain both of RIF and INH resistance. The resistance mechanism is mediated by missense mutations in rpoB, the gene encoding the B subunit of the RNA polymerase (11). In contrast to INH, the mutations associated with RIF-resistant MTB isolates are located in an 81 bp core region of the rpoB gene (12).
2. Objectives
Actually, the major aim of the present study was to detect the statistical information about the distribution of MDR-TB isolates in Lorestan province, in the west region of Iran. In other words, we tried to distinguish the INH-and RIF-resistant MTB isolates by molecular techniques as a rapid diagnostic method. Also, we tried to assay the specific codons in which most mutations occurred.
3. Methods
3.1. Study Population
A total of 106 patients with MTB participated in our study from September 2014 to 2016 in Lorestan province, Iran. Identification of all isolates was done by polymerase chain reaction (PCR) of IS6110.
3.2. Drug Susceptibility Testing
Clinical isolates of MTB were subjected to standard drug susceptibility tests. The proportional method was used to assess the susceptibility of isolates to INH (20 µg/mL) and RIF (40 µg/mL).
3.3. DNA Extraction and PCR
DNA extraction was performed as the following: bacterial colonies from Löwenstein-Jensen medium were collected in a tube containing 100 μL of sterile double-distilled water. Subsequently, an equal volume of chloroform was added and incubated for 20 min at 80ºC. Tubes were centrifuged at 12,000 rpm for 1 min. The supernatant was used as the template for the PCR. katG, rpoB, inhA and IS6110 genes were amplified by PCR using specific primers (Table 1). The PCR mix was the same for each primer pair used. In brief, 5 μL of DNA was added to a final volume of 25 μL of PCR mixture containing 0.3 μL of dNTP, 0.6 μL MgCl2 (50 mM), 0.5μL of primer mix (100 pmol/µL), 2.5 μL of 10X Taq DNA polymerase buffer, and 0.2 μL of Taq DNA polymerase (5U). The thermal conditions were as follows: 5 min at 95ºC that followed by 30 cycles of 1 min at 95ºC, 1 min at appropriate annealing temperature, and 1 min at 72ºC and was also allowed a final elongation step of 5 min at 72ºC. The PCR products were visualized by electrophoresis. DNA size marker (100 bp DNA Ladder, Thermo fisher scientific, USA).
3.4. DNA Sequencing and Analysis
The PCR product of katG, rpoB, inhA genes was used as the template for sequencing, which was performed by Pishgam co., Iran. The DNA sequences were analyzed with BioEdit V7.0.9.
4. Results
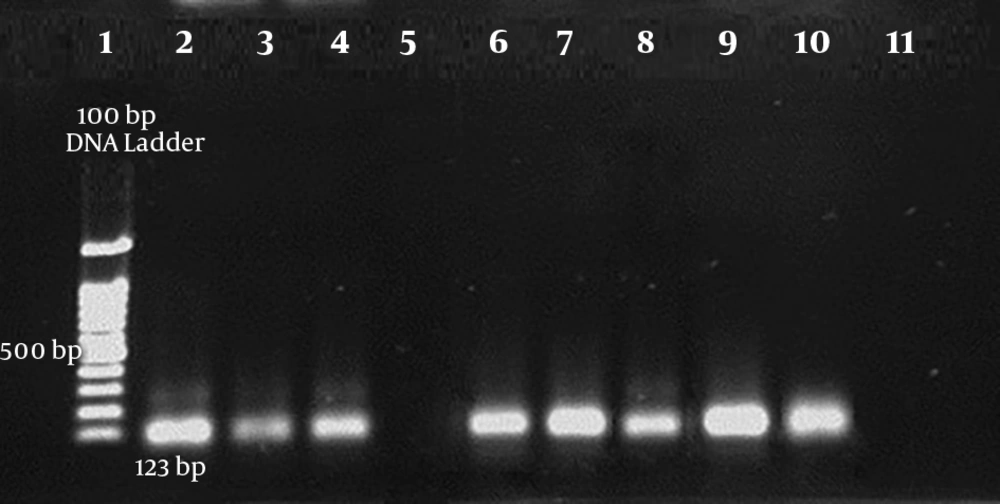
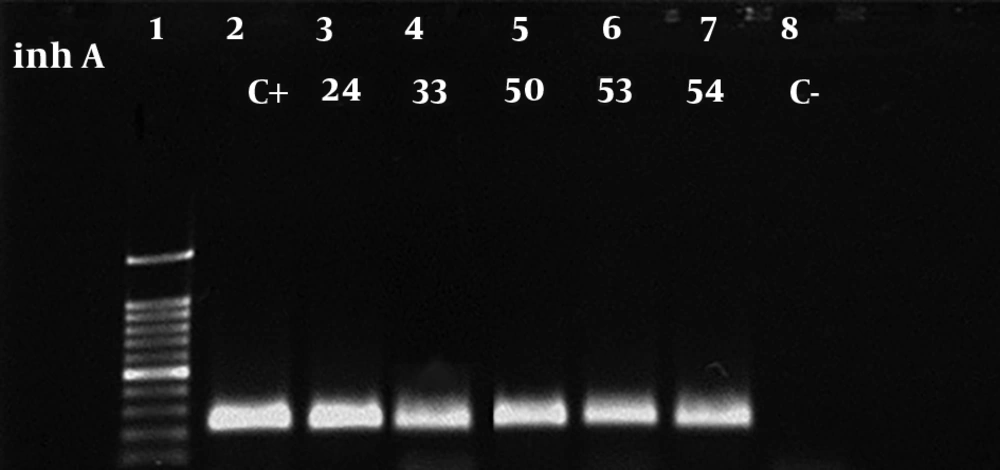
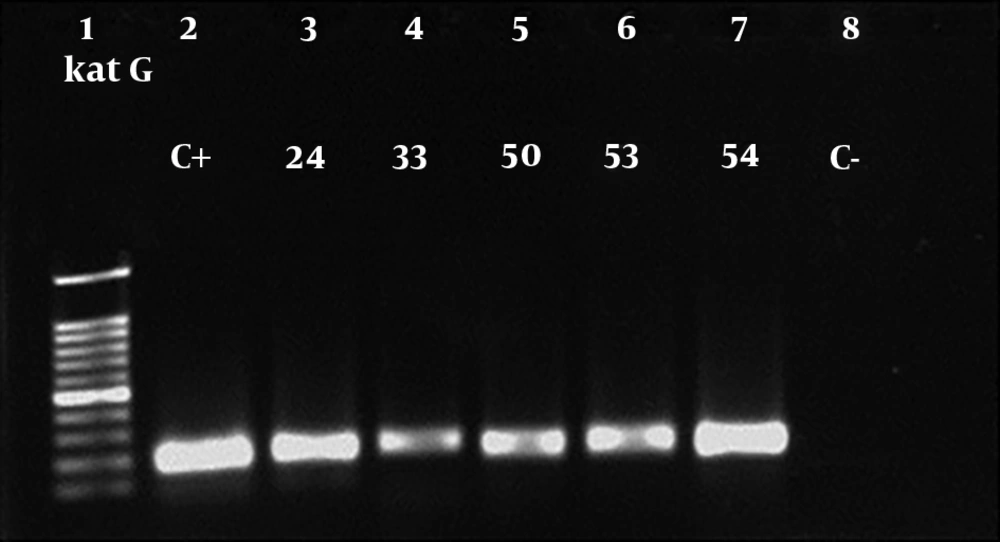
A total of 118 patients were involved in this study, among which 106 cases were identified as MTB based on IS6110-based PCR (Figure 1). Additionally, antibiotic susceptibility testing was performed using the proportional method. The majority of patients belonged to the age group of 15 - 45 in both genders. Table 2 indicates the characteristics of the study population. Eight patients were simultaneously infected with the HIV virus. The results of susceptibility testing of MTB isolates showed that of the total MTB patients, 4 (3.8%), 1 (0.9%), and 1 (0.9%) of whole studied population were resistant to INH + RIF, INH, and RIF, respectively. Thus, 4 (3.8%) isolates were considered to be MDR. Amplification of katG, rpoB, and inhA genes in resistant MTB isolates yielded 209 bp, 157 bp and, 238bp products, respectively (Figures 2-4). A mutation in codon290 of katG gene was detected in 3 out of 4 INH + RIF-resistance isolates, and another isolate had a mutation in codon293 of katG gene (Table 3). One phenotypic INH+RIF-resistance isolate had no mutation in codon290 of katG gene. Also, 4 INH + RIF-resistance isolates had a mutation in codon339 of rpoB gene (Table 3). In 1 out of 4 INH + RIF-resistance isolates, two types of mutations were observed in codon339 and codon347 of rpoB gene. One INH-resistance isolate had a mutation in codon293 of katG gene. A mutation in codon339 of rpoB gene was detected in 1 RIF-resistance isolates (Table 3).
| Variable | Frequency (%) |
|---|---|
| Gender | |
| Female | 37 (34.9) |
| Male | 69 (65.1) |
| BMI (kg/m2) | |
| Normal BMI (BMI ≥ 18.5) | 48 |
| Moderated overweight | 7 |
| Excessive obesity | 1 |
| Malnourished (BMI < 18.5) | 50 |
| History of treatment | |
| Recurrence (with previous treatment) | 11 |
| No previous treatment | 95 |
| Drug resistance | |
| INH + RIF (MDR) | 4 (3.8): 3 co-infection of HIV and MTB, 1 mono-infection of MTB |
| INH | 1 (0.9) |
| RIF | 1 (0.9) |
| No. Isolate | Resistance Pattern | Sequencing Results |
|---|---|---|
| 1 | INH | katG-869 G → C (Thr → Ser) |
| 2 | RMP | rpoB-1016 T → A (Asp → Val) |
| 3 | INH/RMP | katG-No mutation |
| rpoB-1016 T → A (Asp → Val) | ||
| rpoB-1040 T → deletion (X → Leu) | ||
| 4 | INH/RMP | katG-869 G → C (Thr → Ser) |
| rpoB-1016 T → A (Asp → Val) | ||
| 5 | INH/RMP | katG-869 G → C (Thr → Ser) |
| rpoB-1016 T → A (Asp → Val) | ||
| 6 | INH/RMP | katG-869 G → C (Thr → Ser) |
| katG-718 C → deletion (Leu → Miss) | ||
| rpoB-1016 T → A (Asp → Val) |
5. Discussion
Currently, drug resistance is the main problem in the battle against tuberculosis. During the last years, the resistance rate has been steadily increased (15). Identification of MDR-MTB within 6 weeks can provide a phenotypic drug susceptibility testing result and many transmission events can occur during this time (16). The present study was performed according to standard laboratory guidelines and showed a 3.8% prevalence for MDR-MTB. In this study, the highest number of MTB was seen in the men aged 15 to 45 that can be attributed to the further communication of this group with the community. The WHO reported that 67.2% of the global tuberculosis prevalence occurs in males compared to females (17). One well-known symptom of tuberculosis is weight loss and the highest number of MTB was found in people with body mass index (BMI) less than 18.5 (18). Nevertheless, the association between BMI and tuberculosis infection has not been comprehensively understood (19). Most cases of resistant tuberculosis were isolated in Khorramabad, Azna and Kohdasht cities; these communities have the most marginal population of the province. Based on the results of this study, the respondents had basic information about the communal symptoms of tuberculosis and their transmission pathways, which is consistent with previous studies in a rural community in Iran (18), Ethiopia, and Iraq (20, 21). A lack of knowledge of the etiology of the disease can negatively affect the attitudes of the patients towards health-promoting behaviors and preventive methods, since most people with such beliefs may not visit health centers or consider several traditional alternatives (22). As indicated in the study, the prevalence of MDR-TB was significantly higher than in newly diagnosed cases with tuberculosis. Most importantly, patients with tuberculosis who had a history of anti-tuberculosis treatment were 8.1 times more likely to develop MDR-M.TB infection compared with newly diagnosed cases with tuberculosis. Previously treated patients often represent a very heterogeneous group, except those who relapse after successful treatment, those who return after default, and those who start receiving a re-treatment regimen after experiencing previous treatment failure (23). There is enormous evidence that noted that a history of anti-tuberculosis treatment is one of the main contributing factors in the acquisition of MDR-MTB (24, 25). Among previously treated tuberculosis, MDR generally results from the experience of a single drug that suppresses the growth of bacilli susceptible to this drug but allows the proliferation of pre-existing drug-resistant mutants (26). It is the most common type of resistance to the first-line drugs and can emerge against any anti-tuberculosis agent during chemotherapy. The occurrence of MDR-MTB is also due to the lack of prescription of standard drugs, it likely leads to treatment failure and intensifies drug-resistant strains of the population (27). The results of this study showed 7.5% HIV prevalence in patients with tuberculosis. Of the 8 tuberculosis/HIV isolates, 3 were resistant to both RIF and INH. According to the reports of Tuberculosis Control and Leprosy Department of Ministry of Health and Medical Education in 2014, the HIV prevalence index among patients with tuberculosis was 2.5%. The results showed that two main factors for MDR-TB were HIV co-infection and previously tuberculosis treatment. Other studies similarly reported for MDR-TB, which significantly related to HIV epidemics (25, 28). It is understood that HIV infection dysregulates immunological reactions, which leads to irresistible infection, by opportunistic and drug-resistant strains (25). Most studies have reported the prevalence of MDR-MTB and different drug resistance types the Research Center of Tuberculosis of Masih Daneshvari Hospital and Pasteur Institute of Tehran, Iran. Bahrmand et al. reported the 4% frequency of resistance to INH + RIF among 563 patients who referred to the Pasteur Institute of Iran (18). Shamaei et al. found 10 (2.8%) MTB isolates resistant to RIF + INH (29). In 2015, Tavanaee Sani et al. reported 51 of 1,251 patients with MDR from Shariati Hospital, Mashhad, Iran (30). In this study, the frequency of MTB was similar to other Iranian studies with 3.8% frequency. However, our data are contrary to previous studies, which were reported mutations of the codon315 of the katG gene are associated with INH-resistant MT (31, 32). Sadrnia et al., have shown that among of 87 isolates, 33 INH-resistant isolates had a mutation in Ser315Thr of KatG gene, while 21 susceptible isolates had no mutation in codon315 of katG gene (33). Bostanabad et al., with a study on the frequency, location and type of mutations in katG gene of MTB isolate from Belarus that the most mutations were formed in codons315, 309 and 316 of katG gene (34). In 2009, Abdelaal et al., revealed that of 50 isolates, 23 were resistant to RIF and 26 were resistant to INH. The sequencing results showed that 89% of the RIF-resistant isolates had a mutation in the rpoB gene and 92% INH-resistant isolates had a mutation in the katG gene (35). However, mutations in another region of rpoB gene should be studied. It is necessary to check all katG, inhA and kasA gene mutations in INH-resistant isolates. The appropriate design of molecular techniques is required to identify INH + RIF-resistant isolates that had no mutations in the common areas of rpoB and katG genes. The prevalence of drug-resistant tuberculosis in this study, as associated with the WHO reports and national averages, indicates the requirement for management and control of drug-resistant tuberculosis. Although the fact that PCR is a quick method for the detection of MTB resistant strains, it is desirable to use both conventional and molecular methods to obtain more accurate information concerning the resistance pattern.