1. Background
Enterococci are Gram-positive bacteria with low GC content, belonging to the Enterococcaceae family. They have emerged as one of the leading causes of serious nosocomial infections in recent years. Although many species have been identified in the Enterococcus genus, the most common agents associated with clinical complications are E. faecalis and E. faecium (1-3). The emergence of multidrug-resistant enterococci as one of the most frequently encountered nosocomial bacteria has created an increasing health concern worldwide. Therefore, the reliable identification of these pathogens using an easy, fast method is important (4, 5).
Although the traditional culture methods are mostly used for the detection of clinical enterococcal isolates, these methods are time-consuming and dependent on the presence of live organisms in the specimen. As more rapid and sensitive methods, several polymerase chain reaction (PCR)-based procedures have been developed for the detection of Enterococcus spp. The molecular approaches are dependent upon the amplification of DNA and are not affected by low microbial loads (6-8).
Loop-mediated isothermal amplification (LAMP) assay has been recently introduced as a rapid, accurate, and cost-effective diagnostic method for infectious diseases with the potential to overcome the limitations of culture and PCR methods (9, 10). In this technique, the target DNA is amplified under isothermal conditions and four specific primers (F3, B3, FIP, and BIP) are annealed to six separate regions within the target sequence. The use of these primers is associated with high efficiency and specificity (11, 12). Moreover, by using additional loop primers (LF and LB) designed to anneal the loop structure in LAMP, the LAMP reaction can be accelerated, resulting in the reduced amplification time. In the LAMP assay, the white precipitate of magnesium pyrophosphate produced during the reaction can be detected by the naked eye without any additional processing (10, 13).
2. Objectives
The identification methods for classical Enterococcus species by phenotypic methods are often time-consuming; thus, it seems to be a need for developing rapid and specific assays to overcome the problem. In the present study, the LAMP assay was developed as a confirmatory tool for the identification of clinical E. faecalis and E. faecium isolates.
3. Methods
3.1. Bacterial Isolates and Phenotypic Identification
This cross-sectional study was conducted on 57 non-repetitive clinical enterococcal isolates from UTI (urinary tract infection) patients (with urine culture yield of ≥ 105 CFU/mL) in the Department of Urology, Sina Hospital, Tehran University of Medical Sciences, Tehran, Iran. All the isolates were presumptively identified as enterococci by primary phenotypic identification methods and then included in the study. Duplicate isolates from the same patients were excluded (only the first isolate from each patient was included). The isolates were collected over a period of nine months from May 2015 to January 2016. The Review Board and Ethics Committee of Tehran University of Medical Sciences approved the study.
Enterococcus spp. were identified using the standard biochemical and microbiological methods including Gram’s staining, catalase test, motility test, pigment test, Enterococcus selective media (Bile-esculin-azide agar and Slanetz and Bartley agar (Merck, Germany)), salt tolerance test (6.5% NaCl (Merck, Germany)), and acid production from 1% (w/v) sorbitol, sorbose, L-arabinose, D-ribose, sucrose, and raffinose (Merck, Germany) (3).
3.2. Primer Design and DNA Extraction
The LAMP primers for the detection of E. faecium were designed by targeting the efmC gene using Primer explorer V4 software (https://primerexplorer.jp/lamp4). The sequences are shown in Table 1. PCR was performed with the F3 and B3 primers as forward and reverse primers, to confirm their specificity. Moreover, the E. faecalis mtlf gene was used as the target for the LAMP primer design according to a previous study (14). It is worthy of note that the genomic DNA was extracted from pure cultures using a High-Pure PCR Template Preparation Kit (Roche, Germany) according to the manufacturer’s instruction.
| Bacteria | Primer | Sequencing (5’–3’) | Target Gene | Amplicon Size with F3 + B3 | Ref. |
|---|---|---|---|---|---|
| E. faecalis | F3 | GCCATTCCTCATGGAACAG | mtlf gene | 205 bp | (14) |
| B3 | CCACGTTATCCATATCACTACA | ||||
| FIP | CGGTGCCAAAATTAACACCCT | - | |||
| TTAGTGAAAAAATCAGGAATCTGTG | |||||
| BIP | TGCTACCGTATTATTTGGGATTGC | ||||
| AAAGTGCAATTTGTTGGACTA | |||||
| E. faecium | F3 | ACATGGCAAAAGAAGTAGGT | efmC gene | 208 bp | This study |
| B3 | ACTGCAAAATAATCTTTCCCT | ||||
| FIP | CCTTTTTGATGTCTTCTGGTAAT | - | |||
| GGAAACAATAGATATCCACAGTATCCCG | |||||
| BIP | TACAACGGTTTGAATCTTGAAACAG | ||||
| AAATCTTTTTTAGCCGTTTCCATC | |||||
| LF | ATATTCATGCGGATCTGTTCC | - | |||
| LB | GTAACAGCTGGTTCGATAACT |
3.3. LAMP Reaction
The LAMP reaction for the detection of E. faecalis was carried out according to the method described by Xu et al. (14) with slight modifications. The optimized LAMP reaction mixture (25 μL) consisted of 1X Bst buffer (New England Biolabs, UK), 8 mmol/L of MgSO4 (Sigma-Aldrich, USA), 8 mmol/L of each deoxynucleoside triphosphate (dNTP), 0.6 μmol/L of each outer primer (F3 and B3), 1.8 μmol/L of each inner primer (FIP and BIP), and 2 μL of DNA template. The reaction mixture was heated at 94 °C for 5 min in a Thermoblock heat system, and then cooled on ice. Finally, 8 U Bst DNA polymerase (New England Biolabs, UK) was added. Subsequently, the mixture was incubated at 60ºC for 60 min and then, heated at 80ºC for 3 min to terminate the reaction.
To optimize the LAMP conditions for E. faecium, the reaction mixture was prepared using a total volume of 25 µL containing 1X Bst Buffer (New England Biolabs, UK), different ratios of outer, inner, and loop primers, different concentrations of dNTPs (4, 6, 8, and 10 mM), different concentrations of MgSO4 (2, 4, 6, 8, 10, and 12 mM) , 8 U Bst DNA polymerase large fragments (New England Biolabs, UK), and 2 μL of DNA template. The reaction mixture was incubated in a Thermoblock system at various temperatures (63ºC and 65ºC) for various durations (30, 60, 90, and 120 min).
The final amplification LAMP products were visualized by using 3% agarose gel electrophoresis (Merck, Germany) or observing turbidity derived from magnesium pyrophosphate formed during the amplification process (Figure 1). It is worthy of note that the LAMP assays for the efmC and mtlf genes were optimized using E. faecium (ATCC® 19434TM) and E. faecalis (ATCC® 29212TM), respectively, which were later applied to screen the clinical isolates of enterococci.
4. Results
4.1. Phenotypic Identification
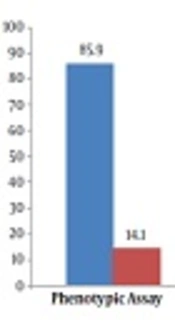
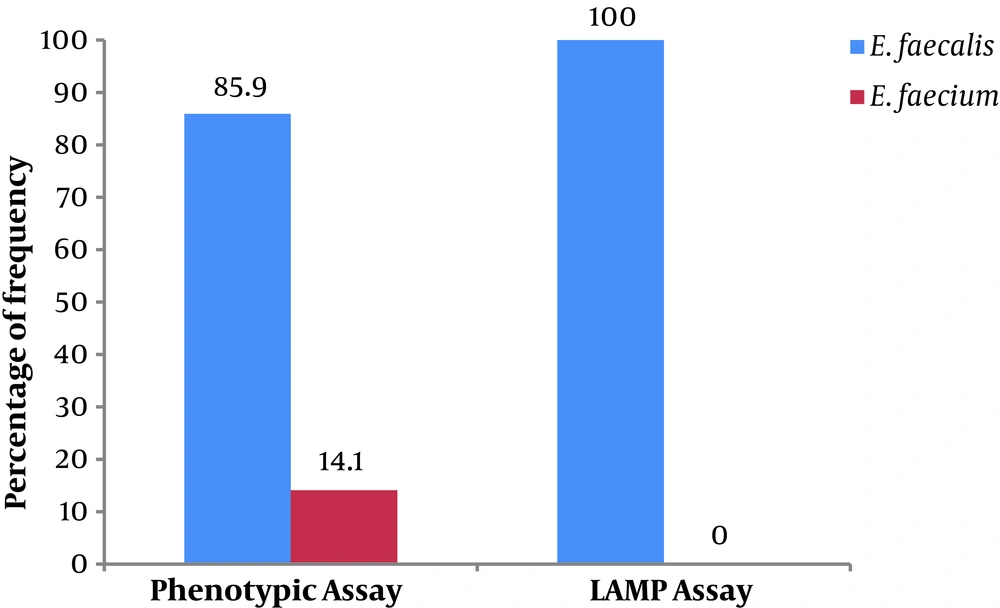
In the present study, we investigated 57 clinical enterococcal isolates from UTI patients. The results of phenotypic testing indicated that of the 57 enterococcal isolates, 85.9% (49 isolates) were E. faecalis and 14.1% (eight isolates) were recognized as E. faecium.
4.2. LAMP Assay
To optimize the LAMP conditions for the detection of E. faecium, we used the pure DNA of E. faecium (ATCC® 19434TM). The ideal ratio of the outer, inner, and loop primers was 1:3:1 (corresponding to 0.6 μmol/L each of F3 and B3, 1.8 μmol/L each of FIP and BIP, and 0.6 μmol/L each of LF and LB). The extracted DNA from E. faecium was achieved optimally at 61ºC. Therefore, the best temperature for the LAMP assay was established at 61ºC, which was used for all subsequent applications. The best time for the amplification of genes in the LAMP reaction was 60 min. Different MgSO4 and dNTPs concentrations were evaluated and the best results were obtained at 6 mmol/L and 8 mmol/L, respectively.
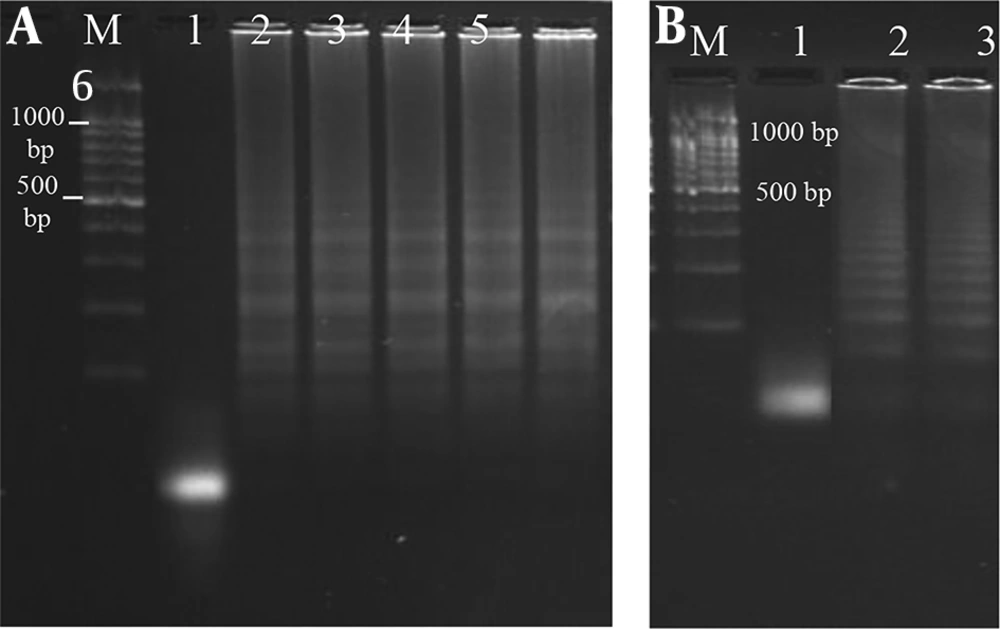
In our study, the LAMP assay was applied for the detection of clinical enterococcal isolates (Figure 2). The results showed that all the 57 enterococcal isolates were identified as E. faecalis (Figure 3).
Electrophoretic analysis of LAMP products. (A) LAMP detection of E. faecalis isolates by targeting the mtlf gene. Lane M, DNA ladder (100 bp); lane 1, negative control (no template); lane 2 - 6, clinical E. faecalis isolates. (B) LAMP detection of E. faecium isolates by targeting the efmC gene. Lane M, DNA ladder (100 bp); lane 1, negative control, lane 2 and 3, E. faecium (ATCC® 19434TM).
5. Discussion
Accuracy in the identification of enterococci to the species level is important for adequate treatment, surveillance, and infection control. Phenotype testing that involves bacterial growth in selective media has been widely used to diagnose enterococcal isolates (15, 16). However, these methods are time-consuming and failure in the identification of some Enterococcus species may have implications in clinical practices. Despite great sensitivity, PCR-based methods are not widely used as routine diagnostic tools due to the need for thermal cyclers and highly skilled operators. Therefore, the development of a more simple, rapid, and sensitive diagnostic method is important for the detection of enterococcal isolates (14, 15).
Since its advent in 2000, the LAMP assay has been frequently used for the detection of many organisms such as bacteria because it is user-friendly and has high sensitivity and specificity (17, 18). In the present study, we developed a LAMP assay for the detection of E. faecium and E. faecalis as two important Enterococcus spp. There are some publications about the LAMP assay and its use in the detection of enterococci, but different results have been obtained about LAMP conditions such as reaction temperature, MgSO4, dNTPs, and primers concentrations. In our study, the optimal temperatures were 60 and 61ºC for the detection of E. faecalis and E. faecium, respectively, in the LAMP assay. However, a recent study showed no difference in gene amplification conditions between Gram-negative and Gram-positive bacteria in the LAMP assay (19). In another study, the optimum temperature was 65ºC for gene amplification in the LAMP test for the detection of E. faecalis (6). The optimum conditions for gene amplification in the LAMP assay seem to be associated with polymerase enzymes used in reactions so that the best temperature is dependent upon the activity of Bsm and Bst polymerases as the two enzymes used in LAMP reactions.
In the current study, 57 clinical enterococcal isolates were recognized by phenotypic methods as E. faecalis (49 isolates) and E. faecium (eight isolates). The phenotypic prevalence of enterococci in this study is consistent with reports from many studies indicating Enterococcus faecalis as the most common cause of enterococcal infections (20, 21).
Furthermore, in this study, the LAMP assays targeting the mtlf and efmC genes were used to confirm E. faecalis and E. faecium, respectively. The results from the blasting of sequences showed that the selected sequences for the specific detection of the abovementioned species were highly specific. Moreover, conventional PCR was performed using external primers F3 and B3 to confirm the validity of the LAMP assay results. The LAMP assays showed that all the 57 enterococcal isolates were identified as E. faecalis. Therefore, the results indicated the importance of using the LAMP assay for the rapid and reliable identification of infectious agents. The results of the current study are in line with the results of other studies. Velasco et al. showed no correlation between phenotypic techniques and PCR-based genotypic methods for the identification of Enterococcus spp. In this study, up to 15.8% of the strains of E. faecium were misidentified based on phenotypic methods (15). Another study indicated that PCR-based techniques are more effective than biochemical methods for the complete identification of enterococcal isolates (22). Overall, the studies emphasize the inadequacy of phenotypic methods alone for the correct identification of Enterococcus species, particularly E. faecium. It is noteworthy that although the LAMP assay is a more rapid and simpler method than conventional PCR, the feasibility and cost-effectiveness of molecular diagnostic methods for patients should be considered.
One of the limitations of this study is that the LAMP assay was used only for the identification of E. faecalis and E. faecium as the most common agents associated with clinical complications although the LAMP method can be accomplished for the differentiation of all Enterococcus species. Furthermore, in the present study, the LAMP assay was developed as a confirmatory tool for species identification of cultured clinical isolates while this diagnostic method can rapidly identify infectious agents in clinical specimens.
5.1. Conclusions
The study highlights the importance of PCR-based genotypic techniques in routine use for the identification of Enterococcus spp. Although most laboratories employ phenotypic methods for bacterial identification, this study provides strong evidence that a number of enterococcal isolates may be misidentified when only biochemical tests are used. Considering the rapid performance of the LAMP assay, it may have implications as a diagnostic and confirmatory tool for the identification of clinical Enterococcus species.