1. Background
Acinetobacter is an obligate Gram-negative and aerobic coccobacillus, which has been among the most important bacterial infections in the last three decades (1). The most common problem with the treatment of infections caused by this bacterium is the resistance to most antibiotics (2). Antibiotic resistance capacity can be transmitted among bacteria, leading to an increasing resistance day by day (3). Acinetobacter baumannii is mainly responsible for the most antibiotic-resistant cases in this genus in which the multidrug resistance has led to many problems in the treatment of hospital infections both in the costs of treatment and in the recovery of patients (4-6). A few studies have been done to investigate the virulence factors of this bacterium; however, the most important ones that contribute to the pathogenesis of A. baumannii include the ability of biofilm formation, the presence of polysaccharide in the cell wall, phospholipases, iron absorption capability, external membrane vesicles, OmpA outer membrane proteins, and penicillin-binding protein (7, 8).
It has been established that A. baumannii can apparently survive on artificial surfaces for a long period; thereby, allowing it to persist in the hospital environment due to its ability to form biofilms (9). Biofilms provide bacteria with three main advantages, including trapping the essential elements and nutrients for bacterial usage, tolerate hard conditions as well as the protection against the host’s immune system, and the opportunity to transfer antibiotic resistance genes (10). Furthermore, the availability of fewer nutrients in deeper space within the biofilm and consequently a slower metabolism can prevent the bacteria from taking up an antibiotic and antimicrobial efficacy (11, 12). The process of biofilm formation in many bacteria is mediated by flagella; however, for A. baumannii, pili seem to be involved in this process under the regulation of CsuA/BABCDE operon (13). It has been shown the inactivation of CsuE results in the prevention of pili production and the formation of biofilms. This indicates that the CsuA/BABCDE system plays a prominent role in the initial stage of biofilm formation (14).
Owing to the lack of new synthetic antimicrobials to treat MDR Gram-negative infections, attention should be increasingly focused on natural compounds either as stand-alone or adjunctive therapies. Curcumin is a polyphenol mixture extracted from the yellow root of turmeric, which has several pharmacological effects including anti-inflammatory, anti-oxidant, anti-cancer, anti-diabetic, anti-allergic, and antibacterial activity (15). Unfortunately, curcumin faces problems such as low solubility, chemical and biological instability. Today, the use of nanotechnology has improved the biocompatibility of curcumin and the slowdown of high decomposition. On the other hand, the increase of its stability in the blood and the reduction of its toxic effects have been widely investigated (16).
2. Objectives
Regarding the effect of Curcumin on biofilm in various bacteria such as A. baumannii, this study was aimed to investigate the anti-biofilm activity of curcumin nanoparticles and its influence on Biofilm-related gene expression in A. baumannii in vitro.
3. Methods
3.1. Study Samples and Subjects
This descriptive-cross sectional research was conducted from February 2016 to December 2017. Seventy clinical specimens were included for investigation. The sample size calculation formula was:
With assumption of a confidence level of 95%, the power of 80%, and a precision of 10% of the sample size for a variety of strains. Various clinical specimens included tracheal, sputum, urine, burns, wounds, blood, and unknown resources were collected from patients referring to Firoozgar Hospital (Tehran, Iran).
3.2. Bacterial Culture
Different cultural and biochemical tests were used for the identification and isolation of the Acinetobacter species. Seventy A. baumannii isolates were studied. All isolates were stored at -70°C in microbank vials (Thermofisher, UK) and thawed prior to use.
3.3. Determination of Antibiotic Susceptibility by Disc Diffusion Method
Sensitivity of A. baumannii isolates to eleven antibiotics of different classes, including Piperacillin, Ceftriaxone, Cefepime, Imipenem, Polymyxin, Gentamycin, Tetracycline, Ciprofloxacin, Trimethoprim, sulfamethoxazole, Ticarcillin clavulanic prepared from Rosco company (USA) was assessed by disk diffusion method in a Mueller Hinton Agar (MCC) based on the CLSI instruction. The strains of Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as the reference strains to control antibiogram discs. Ultimately, resistant strains were grouped in one of the following categories: (a) Carbapenem-resistant, (b) MDR (multidrug-resistant), (c) XDR (extreme drug-resistant), and (d) PDR (pan drug-resistant) (17).
3.4. Biofilm Formation Assay
This assay was performed according to a previously reported protocol, with some modifications (18). The level (optical density; OD) of crystal violet present in the destaining solution was measured at 570 nm using a microtiter plate reader (SeikagakuCo., Tokyo, Japan). Each assay was performed in triplicate and the mean OD570 value of the tested wells was applied to determine biofilm production ability. As a control, an uninoculated medium was used for the calibration of OD. As it is indicated in Table 1, the results of biofilm formation were classified as biofilm-negative (-), weak (+), medium (++), and strong (+++). In this study, the standard strain of E. coli ATCC 25922 was used as a positive standard control.
| Biofilm Class | Status | Results |
|---|---|---|
| If OD ≤ ODc | Non-adherent | - |
| If ODc < OD ≤ 2 × ODc | Weakly adherent | + |
| If 2 × ODc < OD ≤ 4 × ODc | Moderately adherent | ++ |
| If 4 × ODc < OD | Strongly adherent | +++ |
3.5. Determination of Nanocurcumin Minimum Inhibitory Concentration
Minimum inhibitory concentrations (MICs) were determined in corning 96-well microtiter plates (Corning, Amsterdam, The Netherlands). MIC of nanocurcumin was determined against all 25 isolates of A. baumannii to nanocurcumin, the strong biofilm producer, according to the British Society of Anti-Microbial Chemotherapy (BSAC) susceptibility testing guidelines (19). Based on a previously reported instruction, nanoparticles of curcumin (nanocurcumin) were prepared by a process based on a wet-milling technique and were found to have a narrow particle size distribution in the range of 2 - 40 nm (20). Required volumes of curcumin stock solution were added to Mueller-Hinton agar (MHA) to achieve the final concentrations of 8, 16, 32, 64, 128, 256, and 512 µg/L. Equal volumes of A. Baumannii 105 CFU) in Iso-Sensitest broth were added to each well. After incubation at 37°C for 24 hours in air, wells were checked for turbidity and the MIC recorded as the lowest concentration where no bacterial growth was observed. All microtiter assays were performed in triplicate and mean values were presented. PAO1 strain of P. aeruginosa was used as a positive control strain. A dilution before MIC (sub-MIC) was used for real-time qPCR.
3.6. Quantitative qPCR
RNA was extracted from strong biofilm-producing strains treated with Sub-MIC of curcumin (64 µg/mL) by high pure RNA isolation kit (Roche, Switzerland), according to the manufacturer’s instructions. RNA quantity and quality was measured by nanodrop and the RNA integrity was assessed by gel electrophoresis. For cDNA synthesis, 1 µL of the reverse transcriptase enzyme (4 U/µL), 2 µL of 10X RT-PCR buffer, 1 µL of mixer master, 11 µL of distilled water, and finally 5 µL of the RNA were mixed to reach a final volume of 21 µL. The reaction mixtures were then incubated for 31 minutes at 39°C. Expression assay of CsuE gene was performed by using SYBR Green real-time qPCR in Rotor gene 2000 (Qiagen, USA) using SYBR® Premix Ex Taq™ II mastermix (Clontech, Takara, Japan) according to the following program: holding at 95°C for 10 minutes, and 40 cycles of 95°C for 20 seconds, 58°C for 20 seconds, and 72°C for 20 seconds. Moreover, 16s rRNA and a reaction without cDNA were used as the reference and negative controls, respectively. The expression of CsuE in the treated samples was calculated in comparison to untreated samples. The primer sequences for CsuE gene were forward: CATCTTCTATTTCGGTCCC and reverse: CGGTCTGAGCATTGGTAA and the primer sequences for 16 seconds rRNA were forward: CAGCTCGTGTCGTGAGAT and reverse: CGTAAGGGCCATGATGACTT. A melt curve analysis was also performed after any reaction to confirm specific amplification.
3.7. Statistical Analysis
Data analysis was performed using statistical package for the social sciences (SPSS) Software version 16.0 (SPSS Inc., Chicago, IL, USA). Statistical significance was assessed via the Pearson χ2 test or the Fisher exact test for categorical variables and Student t-test or Mann-Whitney U test for continuous variables.
4. Results
4.1. Specimens and Bacterial Culture
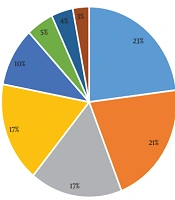
In this descriptive study, 91 samples were successfully taken from patients at three to seven days after admission in Firoozgar Hospital (Tehran, Iran). Seventy isolates of A. baumannii bacteria were successfully obtained, of which 38 isolates had a female source and 32 isolates had a male source. These samples were obtained from the age group of 2 - 75 years. The distribution of A. baumannii isolates collected from different clinical sources are shown in Figure 1.
4.2. Antibiotic Susceptibility
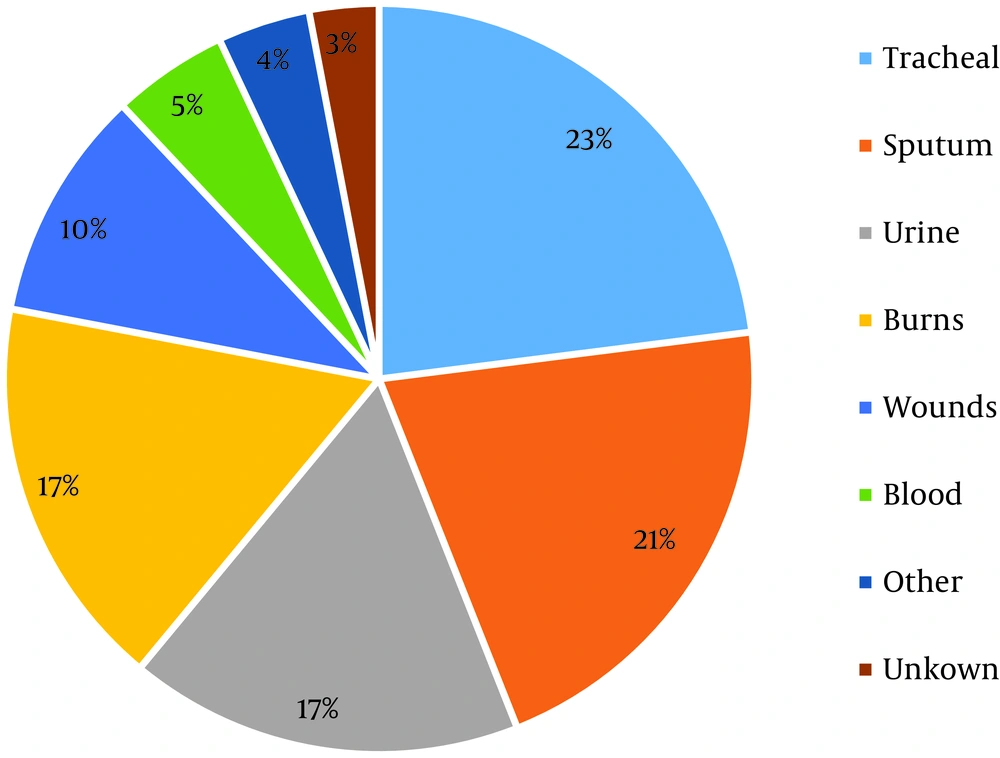
Antimicrobial resistance assay of isolated bacteria showed high resistance to most tested antibiotics, except for polymyxin B (Figure 2). The results also showed that about 20% of the isolates were also sensitive to Tetracycline. These results showed that 92.95% of the A. baumannii isolates were MDR and 86.53% of the isolated were XDR, but there were no PDR-positive isolates.
4.3. Biofilm Formation
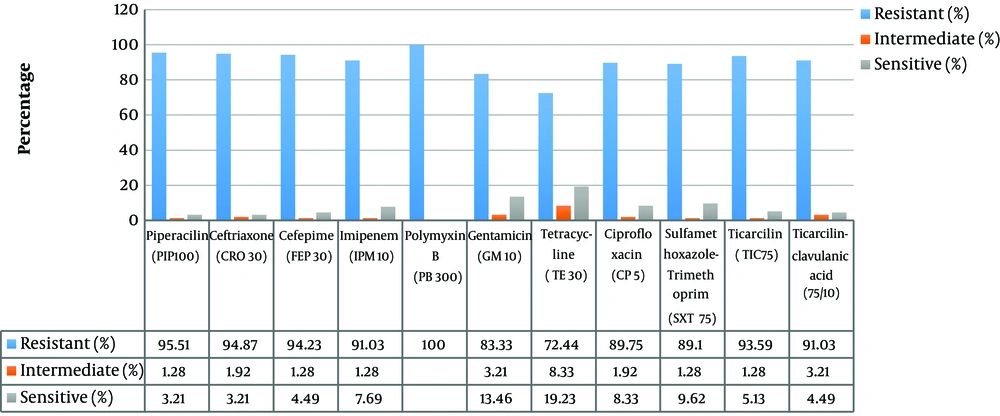
Using a standard qualitative plate microtiter method, the biofilm-producing capacity of 70 isolates was calculated. Two isolates showed no biofilm-producing capacity, 13 isolates were weak, 31 isolates were medium, and 25 isolates were strong to form biofilms (Figure 3). Fifty-two isolates (94.23%) out of a total of 55 medium or strong biofilm-forming isolates were MDR.
4.4. CsuE Expression Results: A Measure of Curcumin Inhibitory Potential
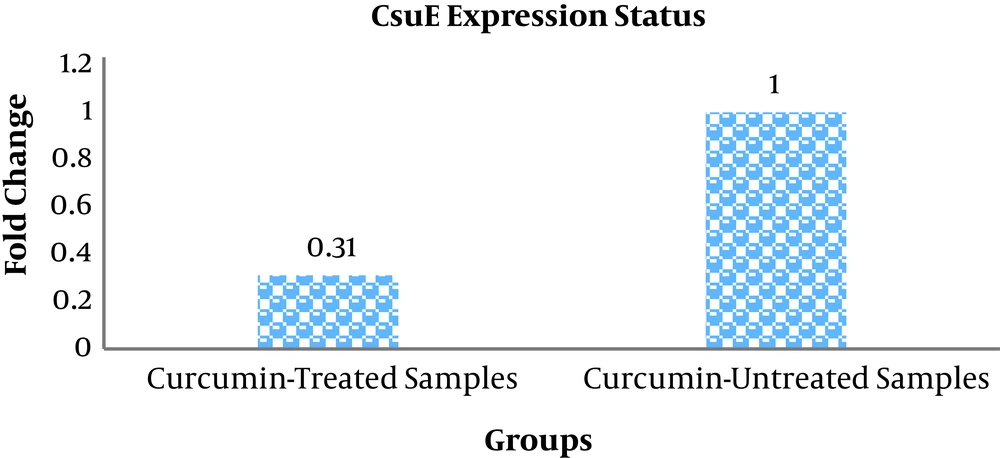
The sensitivity of 25 strong biofilm-producing isolates of A. baumannii was determined based on the minimum inhibitory concentration (MIC) of growth using broth microdilution method to determine the susceptibility to curcumin. The MIC for all of the isolates was 128 µg/mL of the nanoparticle. All of the isolates were treated with a Sub-MIC concentration of 64 µg/mL of curcumin nanoparticles. Twenty-three isolates treated with curcumin (compared with untreated isolates) showed down-regulation and only 2 isolates showed up-regulation in CsuE gene. Totally, the fold change of the CsuE gene, a biofilm-producing-related gene, in the treated samples with curcumin was equal to 0.31 in comparison to the untreated samples (Figure 4).
5. Discussion
Strains belonging to the Acinetobacter baumannii-Acinetobacter calcoaceticus cluster are emerging as problematic opportunistic pathogens due to a rapid increase in multidrug or pan-drug resistance (13). The present study aimed to investigate the effect of curcumin nanoparticles on biofilm-related CsuE gene expression in A. baumannii in vitro. In this study, antibiotic resistance and frequency of MDA and XDR A. baumannii isolates, biofilm formation capacity, as well as the MIC for curcumin nanoparticles were determined. Our data showed that this bacterium had high resistance to most antibiotics, except for polymyxin B. Twenty-five strains showed high biofilm formation capacity. MIC for curcumin was 128 µg/mL in all 25 strains. Expression results indicated that CsuE gene was downregulated to 0.31 fold in curcumin-treated samples compared to untreated samples.
The findings of the current study revealed that all isolates were susceptible to at least one antibiotic, but most isolates were resistant to most antibiotics. Similar to our data, Bassetti and Shakibaie studies showed that different isolates of A. baumannii were resistant to most antibiotics, and 70% of A. baumannii isolates were resistant to 3 or more antibiotics (9, 21). According to various reports, the majority of A. baumannii isolated in Iran were resistant against the first-line drugs of Aminoglycosides, Ceftazidime, Fluoroquinolones, Imipenem, Meropenem (22, 23). MDR-positive A. baumannii isolates have been reported in different studies, wherein, the frequency varies from 40% to 80% in general (24). In the present study, the percentage of MDR-positive A. baumannii isolates was 95.92%. These results indicate that the number of MDR-positive A. baumannii is rapidly increasing in Iran. Therefore, studying the resistance of the A. baumannii, might provide enough information for physicians to select appropriate therapies against infections caused by this organism.
Bacterial biofilms are associated with problems in the prevention and treatment of A. baumannii infection. Curcumin is an unstable, reactive, nonbioavailable compound, while it is an established therapeutic agent and is effective against various strains of Gram-negative and Gram-positive pathogens. Its mode of action at the molecular level has not been established, but it is thought to disrupt bacterial membranes (14). On the other hand, the formation of biofilms has been indicated that can be controlled by coating surfaces with nanoparticles (25). Our findings showed that most of the isolates possess a medium strength of biofilm production. These findings were almost consistent with previous studies in which the percentage of biofilm-producing bacteria was reported as % 63.6 (26), 62% (27), and 60% (28). The findings of this study also showed that there is a significant relationship between the biofilm-producing strength and the resistance to various antibiotics; thus more than 90% of the biofilm-producing bacteria had multidrug resistance properties, which was in accordance with the published reports (26, 28).
In this study, the mean curcumin MICs were determined in 25 strong biofilm-producing isolates and a sub-MIC concentration of 64 µg/mL was used for the treatment of strong biofilm-producing isolates. Of the 25 strong biofilm-producing isolates examined, 23 treated isolates compared with untreated isolates were downregulated in terms of CsuE gene. This can be explained that the addition of the curcumin nanoparticle promotes the decreased expression of CsuE in comparison to untreated samples. Studies have shown that the ability of Acinetobacter strains to form pili and attach to create a biofilm on living surfaces depends on the expression of the CsuE gene (29). Inactivation of CsuE has been shown to lead to the inability of pili production and the formation of biofilms (13). It could be concluded that downregulation of CsuE gene by nanocurcumin highlights the anti-biofilm activity of this compound that can be helpful in decreasing/eliminating of antibiotic resistance by providing new and more effective therapies. There are several limitations to the present work, especially no assessment of antibacterial properties of curcumin by disk diffusion method. Besides, the examination of the effects of other nanoparticles on the formation of biofilm, investigation the toxicity of curcumin nanoparticles, study of the curcumin effects on the animal models, and the evaluation of the curcumin effects on other biofilm-producing-associated genes are suggested for future studies.
5.1. Conclusions
The ability of A. baumannii to form biofilms can lead to a high level of antibiotic resistance and survival in the environment. Considering the inhibitory effect of curcumin nanoparticles against the biofilm-associated CsuE gene, it is suggested to use this nanoparticle as a complementary therapeutic agent against A. baumannii to likely inhibit its virulence gene.