1. Background
Hand, Foot, and Mouth Disease (HFMD) was first discovered in 1957 in New Zealand. Coxsackievirus was purified from Canadian patients in 1959 as an etiologic agent of HFMD. Enterovirus 71 (EV71) is the other cause of HFMD that was first introduced in California (1). HFMD is one of the most common childhood diseases. The symptoms include short-term fever, typical vesicular rashes on the palms and soles, and oropharyngeal sore. In rare cases, the patients may face neurological symptoms such as encephalomyelitis, aseptic meningitis, acute flaccid paralysis, neurologic pulmonary edema, and hemorrhage. Blister (the clear liquid or pus inside the vesicular rash), which is sometimes accompanied by cough, rhinorrhea, and loss of appetite can be seen in some patients. However, these signs need to be confirmed with virus detection by RT-PCR, virus isolation, or other techniques (2). Although the major causes of the disease are EV71 and Coxsackievirus A16 (CVA16), others such as CVA6, CVA4, and CVA10 have also been reported (3-5). The disease can be spread by food and water resources contaminated by wastewater (6). Human Enteroviruses (HEV) belong to the Picornaviridae family, Enterovirus genus. Based on phylogenetic properties, they are divided into four HEV A-D species. HFMD is mostly caused by group A HEV that contains CVA2-8, 10, 12, EV71, etc. (7, 8). Recent studies have shown that most outbreaks of HFMD in countries including Singapore, Finland, and Spain have been due to CVA6 (7). In most cases, HFMD is a self-limited disease but can be dangerous in children with sudden fatal symptoms such as pulmonary hemorrhage four days after skin lesions (4).
2. Objectives
This study aimed at determining the phylogenetic and evolutionary analysis of HFMD etiologic agents in Iran for the first time in an outbreak that occurred on 15th October 2013 in Ahvaz, Southwest Iran.
3. Methods
Sixteen serum samples of HFMD patients comprising 10 (66.6%) boys, five (33.4%) girls, and one unknown patient with a median age of 4.5 ± 3.7 were collected during an outbreak of HFMD in October 2013 from Aboozar Children’s Hospital (the only children’s hospital in the province and Southwest Iran) in Ahvaz, Khuzestan province. All of the samples were taken with patients’ consent (by signing questionnaires). Based on the West Virginia Department of Health and Humans definition, three or more cases of HFMD within one week at the same facility could be defined as an HFMD outbreak (https://oeps.wv.gov/toolkits/documents/hfmd/hfmd-guidelines.pdf). The outbreaks of HFMD are usually benign and self-limited. All children with HFMD symptoms were recognized and confirmed by a pediatrician based on the following inclusion criteria: all or some of the symptoms including fever, rash on palms and soles or buttocks, sore throat, herpangina and blister (the clear liquid or pus inside the vesicular rash), which was at times accompanied by cough, rhinorrhea, and loss of appetite. Most of the cases had a fever, sore throat, and rash on palms and soles. Other non-viral causes of rash such as eczema were excluded from the study by the pediatrician. All the patients had a mild disease without any severe complications like meningitis or encephalitis. Blood samples were taken by nurses from patients in the hospital based on the abovementioned criteria and pediatrician advise. The sera were separated as soon as possible and transferred to the virology department on ice. All the samples were kept at -70ºC for subsequent analysis.
3.1. Genome Extraction and cDNA Synthesis
Viral RNA was extracted by RNX-Plus solution (Sinaclon Bioscience Co., Iran) based on the guanidine thiocyanate method according to the manufacturer’s instructions. Reverse transcription was carried out using a cDNA synthesis kit (Thermo Scientific, USA) briefly as follows: 10 µL of RNA was utilized as a template and 1µL of random primer and 1µL of water were utilized to prepare the first master mix. After incubating the master mix at 65ºC for 5 min and chilling it on ice, the second master mix was prepared based on the manufacturer's instructions. The master mixes were mixed and incubated at 42ºC for 1 h. The reaction was terminated at 70ºC for 5 min.
3.2. Polymerase Chain Reaction for Virus Detection
Each sample was subjected to the Polymerase Chain Reaction (PCR) assay by universal primers for the 5’UTR region for Enterovirus detection. A master mix for the PCR, containing 2.5 µL of 10x PCR buffer mixed with 0.5 µL of dNTP mix (0.2 mM), 0.75 µL (1.5 mM) MgCl2, 10 pM of each primer, 0.2 µL (1 unit) of Taq polymerase, and double distilled water up to 25 µL, was prepared for 35 cycles. The expected 150 and 400 bp bands were visualized by safe stain (Sinaclon, Iran) under UV transilluminator. The first smaller band was utilized for the detection of Enterovirus and the second band was used for 5’UTR sequencing. The PCR primers are shown in Table 1.
| Genomic Region | Sequence | Product Size, bp | Reference | |
|---|---|---|---|---|
| Enteroviruses | 5’UTR | Sense (evf2): TCCTCCGGCCCCTGAATGCG | 150 | (9, 10) |
| Antisense (evr1): ATTGTCACCATAAGCAGCCA | ||||
| 5’UTR | Sense (evf1): CAAGCACTTCTGTTTCCCCGG | 400 | ||
| Antisense (evr2): CACGGACACCCAAAGTA | ||||
| Enterovirus group A | VP3-VP1 | Sense (486): TGGTAICARACIAAITWYGTIGTNCC | 760 | (11) |
| Antisense (488): GTIGGRTAICCITCITARAACCAYTG | ||||
| VP1-2A | Sense (487): ATGTWYGYICCICCIGGIGCNCC | 450 | ||
| Antisense (489): AYIGCICCISWITGYTGNCC | ||||
| Enterovirus group B | VP3-VP1 | Sense (008): GCRTGCAATGAYTTCTCWGT | 650 | (12) |
| Antisense (013): GGIGCRTTICCYTCIGTCCA | ||||
| VP1-2A | Sense (012): ATGTAYGTICCICCIGGIGG | 450 | ||
| Antisense (011): GCICCIGAYTGITGICCRAA |
Enterovirus Universal and Specific Primers
3.3. PCR for Virus Genotyping
In order to determine the virus genotype for groups A and B Enteroviruses, the PCR assay was designed by specific primers for VP3-Vp1 and VP1-2A half of the Enterovirus VP1 region (Table 1). The PCR components and their concentrations were the same as the earlier reactions mentioned for 5’UTR. The expected 650 and 450-bp bands for VP3-VP1 and VP1-2A regions, respectively, for Enterovirus group B and 740 and 450-bp bands for Enterovirus group A were visualized under UV transilluminator. Finally, positive Enterovirus samples for the 5’UTR and VP1 regions were confirmed by DNA sequencing (Bioneer, Korea) while phylogenetic trees were constructed by comparing the sequences.
4. Results
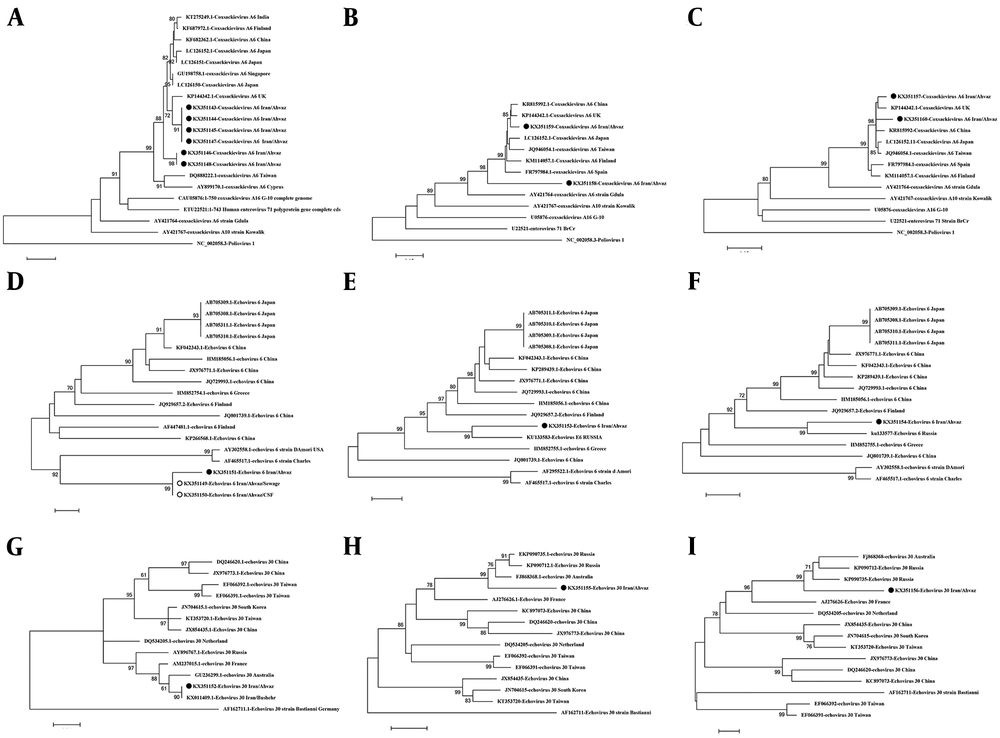
Among the 16 specimens, nine (56.25%) were PCR-positive with the universal primers for the 5’UTR region. Six (66.6%) out of the nine positive cases were from males while two (22.2%) were from females and one positive sample was from an unknown patient (with no information about its gender). The PCR assay with specific primers for the VP1 region was carried out on these nine samples. Nine PCR products of the 5'UTR region, five PCR products of the VP3-VP1 region, and four PCR products of the VP1-2A region of the Enterovirus-positive cases were sequenced (Bioneer Company, South Korea). Seven of the nine samples were recognized as Coxsackievirus A6 while two of them were Echovirus 30 and Echovirus 6. The 5’UTR sequence of the Echovirus 6 sample in our study showed 94% similarity to the wide spectrum of group B Enteroviruses, especially Echovirus 24, 21, and 15, not to a specific serotype; it also showed 93% similarity to Echovirus 6, strain Russia, in its VP1 region. Another sample showed 93% similarity to Coxsackievirus A6 that was lower than other Coxsackieviruses in this study. Phylogenetic trees were drawn for all the sequenced samples based on the 5’UTR, VP3-VP1 half, and VP1-2A half regions (Figure 1).
Phylogenetic tree construction based on different parts of the Enterovirus genome with MEGA 6 software. All the trees were constructed by the NJ method and evaluated by the interior-branch test with 1000 replications. The values under 70% were omitted. A, 5’UTR of Coxsackievirus A6; B, VP1 of Coxsackievirus A6 (VP3-VP1 side); C, VP1 of Coxsackievirus A6 (VP1-2A side); D, 5’UTR of Echovirus 6; E, VP1 of Echovirus 6 (VP3-VP1 side); F, VP1 of Echovirus 6 (VP1-2A); G, 5’UTR of Echovirus 30; H, VP1 of Echovirus 30 (VP3-VP1 side); I, VP1 of Echovirus 30 (VP1-2A side).
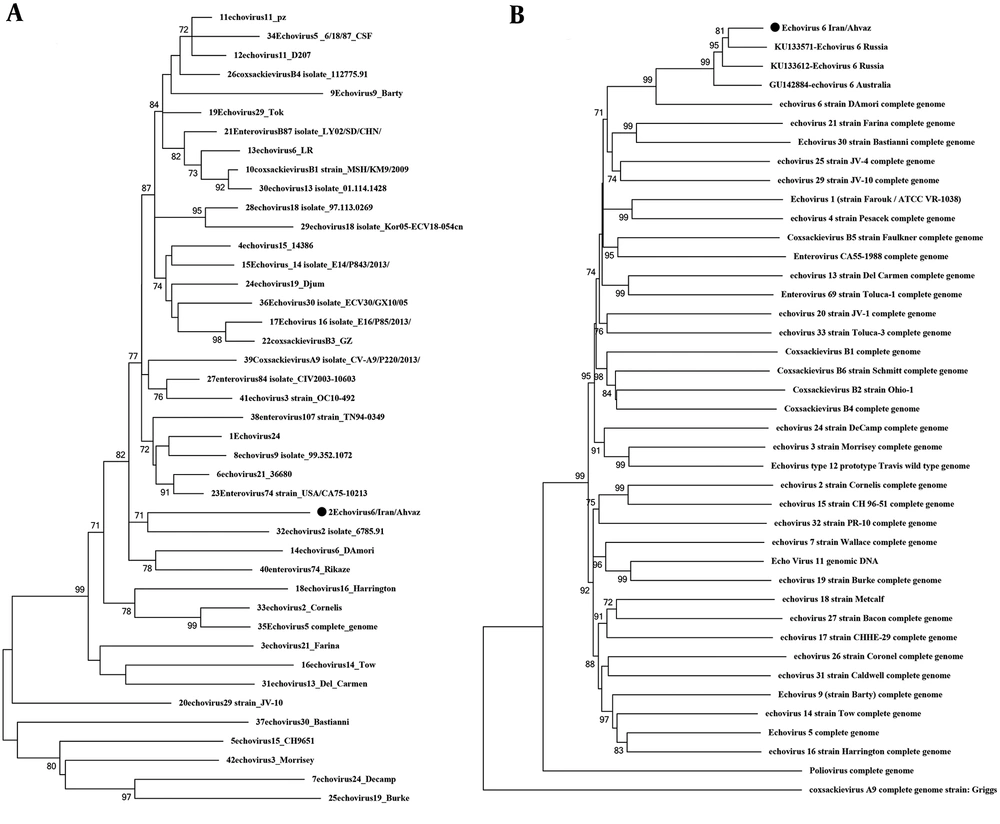
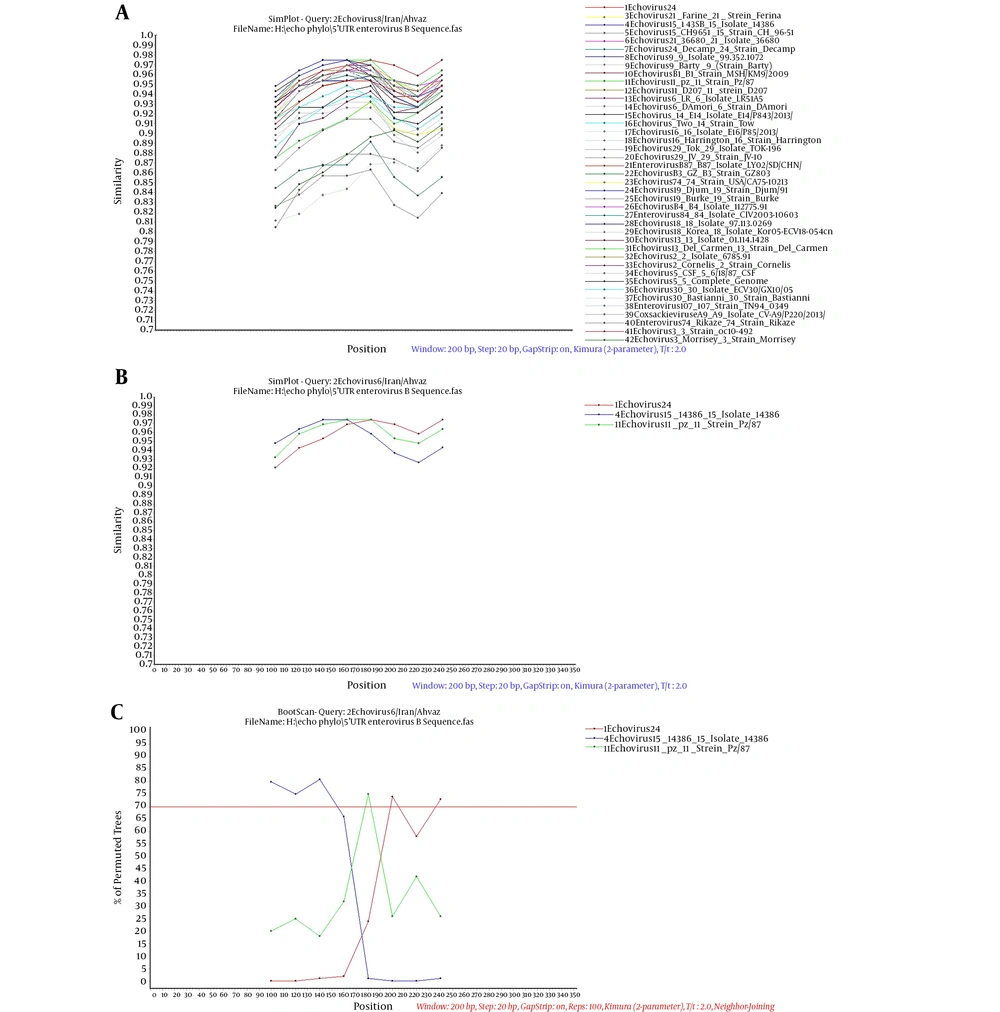
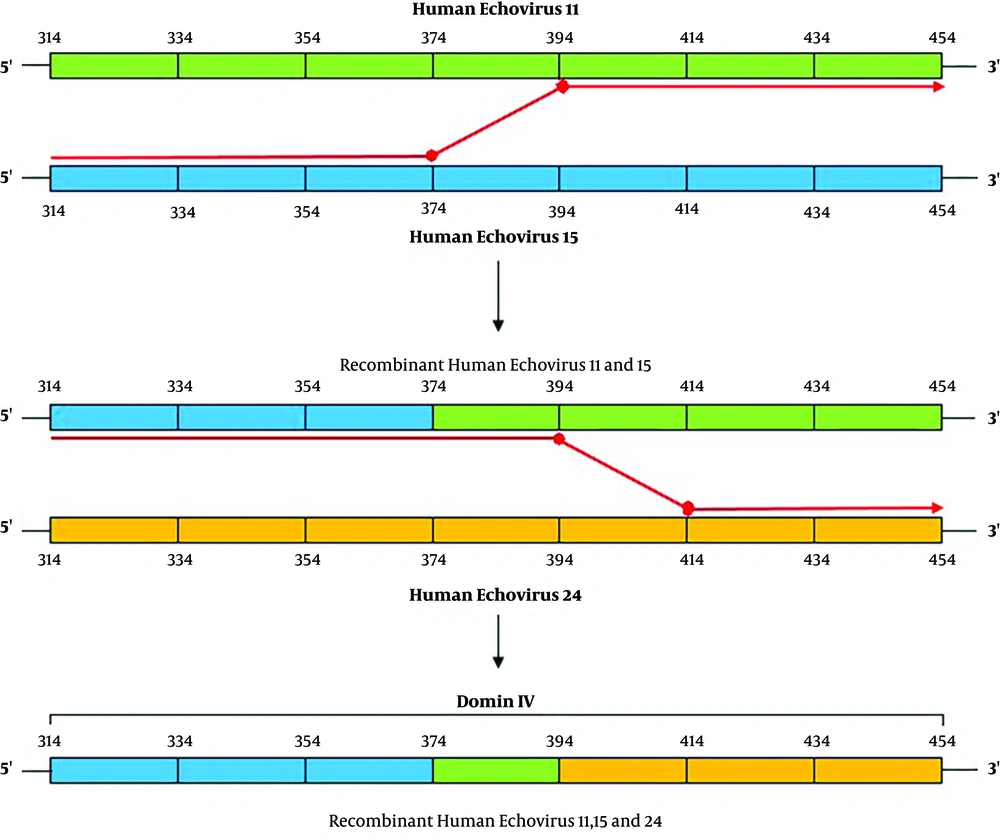
In four cases, CVA6 in its 5’UTR region was in the same cluster as a strain from the UK and in two cases, it was in a separate cluster. The VP1 phylogenetic tree of CVA6 showed the same appearance as the 5’UTR tree. In the 5’UTR tree, Echovirus 6 was in the same branch as strain D, Amore, and Charles (Echovirus 6 prototypes), which was similar to the VP1 phylogenetic tree. Echovirus 30 detected in this study was similar to Bushehr strain that was isolated from Bushehr, South Iran, and created the same cluster as strains from Russia, Australia, and France, which was similar to the Echovirus 30 VP1 tree. A databank of the 5’UTR and VP1 regions of Enterovirus group B prototypes was prepared for comparing Echovirus 6 5’UTR and VP1 trees. These two trees showed different appearances in the arrangement of branches and clusters. This difference could be a sign of recombination (Figure 2). For better evaluation of recombination, Simplot analysis was done and demonstrated the possibility of a recombination event between echovirus 24, 15, and 11 in two nucleotide sites of 5’UTR, positions 374 and 394 (Figures 3 and 4).
Phylogenetic tree construction based on A, 5’UTR and B, VP1 regions of Enterovirus group B genome with MEGA 6 software. The trees were constructed by the NJ method and evaluated by the interior-branch test with 1000 replications. The values under 70% were omitted. In the 5’UTR analysis, Echovirus 6 was put in the same branch as Echovirus 2; however, VP1 analysis was put in the same branch as Echovirus 6 isolate as expected.
A, Simplot analysis of 5’UTR of Echovirus 6 compared to Enterovirus Group B prototypes and isolates. B, many similar isolates to E6 were selected and separated from others for boot scan analysis. C, Boot scan analysis confirmed that three serotypes including E11, E15, and E24 had many similarities to E6 serotype detected in this study. The test was carried out in windows of 200 bp and steps of 20 bp. The threshold of boot scan test was 70%.
5. Discussion
HFMD is a universal disease that usually occurs in children. A member of the Picornaviridae family is recognized as the highly contagious viral agent responsible for HFMD that has been separated in various countries including France, USA, India, Japan, etc. (13, 14). Although the main causes of HFMD are CVA16 and EV71, other serotypes such as CV A6, A10, A4, B3, B5, etc. have also been reported (15). Recent studies have shown the turning of the etiologic agent of HFMD from current serotypes such as EV71 and CV16 to CVA6 as an important agent for HFMD. For instance, the 2012 outbreak of HFMD in Thailand was because of CVA6, but from 2008-2011, CVA16 and EV71 were the major viral agents (16). It is confirmed by similar reports from China, Singapore (2008), Finland (2008), Japan (2010), Taiwan (2010), Thailand (2012), India (2012), and North America (2012) (3, 14, 16-21).
In this study, we examined 16 serum samples collected from an outbreak of HFMD in October 2013 in Ahvaz, Southwest Iran. The predominant virus was CVA6 but E30 and E6 also were identified in two cases in our study. Although group A Enteroviruses are mostly reported in HFMD, in some rare cases, Echoviruses as the members of group B Enteroviruses can cause HFMD. For example, in China’s outbreak in 2003, numerous cases of E19 and E30 were isolated with most of the HFMD patients suffering myocarditis and aseptic meningitis. Other echoviruses such as E6, E11, E24, and E25 were reported, as well (22-25). The remarkable point is that we previously detected Echovirus 6 in the Cerebrospinal Fluid (CSF) of a patient with aseptic meningitis (four years ago) and from wastewater (two years ago) in our lab in Ahvaz (Figure 1E). It seems that there is a circulation of Echovirus 6 in the environment and community. The results of a similar study in Greece proved the presence of two isolates of E6 in wastewater in 2006 that were responsible for an aseptic meningitis outbreak in 2007 (26). Based on Thoelen et al.’s report, VP1 diversity in 33 sub-genotypes of Echovirus 6 was between 0.3% and 3.7% in sub-genotype B and between 0.0% and 6.5% in sub-genotype A (27). In our study, we observed 93% identity in the VP1 region in the BLAST result of Echovirus 6, which indicated 7% diversity in the VP1 region and could be considered as a new sub-genotype. There was also a CVA6 isolate with 93% identity, which created a separate branch in the VP1 phylogenetic tree (Figure 1B).
There are two important ways of diversity in the viral RNA: I) the high mutation rate (one mutation per 1000 to 10000 nucleotides) that leads to creating a similar community of viruses with related sequences called quasispecies and II) the recombination event in nonstructural regions such as 5’UTR, P2, and P3 (25). For more investigation, the phylogenetic trees of 5’UTR and VP1 (first half) were drawn. As an interesting result, the two trees showed different appearances in the arrangement of branches and clusters. In the 5’UTR tree, Echovirus 6 was put in the same cluster as Echovirus 2 but in the VP1 tree, as expected, it was put in the same cluster as Echovirus 6 strain D and Amore as the prototypes of Echovirus 6. The study of Abid et al. showed that the difference in the appearance of phylogenetic trees (arrangement of branches and clusters) in the VP1 region compared to the 5’UTR region is a sign of recombination (28). Oberste et al. and Zhang et al. also reported that a recombination event can be observed between different serotypes of Enterovirus group B based on differences in the appearance of trees in the structural and nonstructural regions (29, 30). The evidence shows the possibility of recombination in the 5’UTR region of E6 in this study. To prove this hypothesis, a databank of 5’UTR sequences of different prototypes of Enterovirus group B and some isolates that were similar to our sequences in the BLAST result were constructed and investigated by Simplot software to discover the signs of recombination. Therefore, 42 5’UTR sequences of Enterovirus group B prototypes and sequences from 5’UTR BLAST results were compared with E6 5'UTR in this study. Echovirus 24, 15, and 11 showed more similarities with E6 sequence and a possible recombination event was observed in two points. Simplot analysis was proved by the boot scan test (Figure 3C).
There are two possible ways of recombination in Enteroviruses: I) the replicative copy choice mechanism that is a “template change” process by RNA polymerase during negative-strand synthesis and II) the non-replicative mechanism that is based on the breakage of RNA template and ligates with different segments of the RNA genome (25, 31-33). This recombination can occur in the intertypic or intratypic manners and mostly takes place in nonstructural regions (27, 34, 35). In this study, we could not find the exact type of recombination but Figure 4 shows schematically a hypothesis on the possible way of recombination between Echovirus 24, 15, and 11 in this study based on the replicative copy choice mechanism.
5.1. Conclusions
Molecular characterization of the viral etiologic agent of HFMD in children for the first time in our region showed Coxsackievirus A6, Echovirus 6, and Echovirus 30 as the etiologic agents of HFMD. The rare event of co-circulation of Enterovirus group B (Echovirus 30 and 6) and Coxsackievirus A6 was shown in this study. Based on this co-circulation and the evidence of recombination in Echovirus 6, we suggest that common hygiene practices and sanitation be improved to avoid the circulation of this isolate in community and the advent of new strains.