1. Background
Rabies is a zoonotic, fatal and progressive neurological infection caused by the neurotropic negative-stranded RNA virus from the Lyssavirus genus, family Rhabdoviridae and Mononegavirale order. The infection largely transmitted to humans, mainly by the bite of an infected animal, usually dogs (1). The rabies virus (RV) is not persistent outside of the host and can be inactivated easily by sunlight, heat, and desiccation (2). The infection is characterized by the appearance of severe nervous symptoms that results in paralysis and death (3). Every year, over 60,000 deaths occur from rabies infection. According to a number of investigations, the disease is prevalent throughout the world and endemic in many countries, especially in Asia (56%) and Africa (44%) (4, 5).
The RV genome is approximately 12 kb, which encodes five structural and non-structural proteins in the order (3’ to 5’) nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and RNA-dependent RNA polymerase (L) (6, 7). The RV genome is encapsidated by N, P, and L proteins, which forms a ribonucleoprotein complex that helps in the multiplication of the virus in the cytoplasm of host cells. The G protein is a viral protein exposed on the surface of the RV and is responsible for the viral pathogenicity, and induces protective immune responses against rabies (6, 8).
The remedial approach for rabies infection in both animals and humans is an important issue in medicine because it is re-emerging and becoming a major public health problem all over the world, especially in developing countries. In fact, it can be said the development of effective therapy may depend on a better understanding of the basic mechanisms underlying the pathogenesis of RV (9).
However, direct fluorescent antibody (dFA) test is a rapid, cost-effective, and highly sensitive diagnostic method for detection of the rabies antigens in brain tissue, but it is only efficient for postmortem samples (10). Therefore, a rapid and sensitive antemortem diagnostic method is essential for the diagnosis of rabies in suspected samples. Polymerase chain reaction (PCR) seems to be a reliable diagnostic tool for antemortem diagnosis of this virus nucleic acid in the clinical samples (CSF, saliva, skin biopsy, and corneal impression smear) (11).
2. Objectives
The present study aimed to develop and validate two molecular detection methods, including quantitative real time-polymerase chain reaction (qRT-PCR) and heminested reverse transcription (RT)-PCR based on the conserved regions of L gene of the Pasteur virus fixed strain (PV) and compare them with dFA and MIT tests to establish a sensitive and specific molecular assay for rapid detection of rabies infection.
3. Methods
3.1. Ethics Statement
In this study, four suspected of rabies patients with non-specific symptoms entered this study between April 2017 and April 2018 after obtaining the informed verbal consent. Also, all brains were samples remained from routine diagnostic activities at the National Center for Reference and Research on Rabies, Pasteur Institute of Iran and derived from animals suspected of rabies that had died. The center repository has been registered for research purposes in accordance with the license from the Iran Veterinary Organization.
3.2. Virus Isolates and Sample Collection
Rabies lyssavirus isolates 35009 (35009-RV-CVS, accession number. LT839616) (lethal dose 50%, LD50 > 10-8) obtained from the National Center for Reference and Research on Rabies, Pasteur Institute of Iran were included in this study. Also, specimens of the brain tissues preserved in the archive of the center were randomly selected so that the final results of the rabies test were 68% (34 cases) of the total positive population (50 cases). All brain tissue samples were previously diagnosed by dFA and MIT and then coded prior to qRT-PCR and heminested RT-PCR assays. All samples were stored at -70°C.
Moreover, three saliva samples at intervals of three to six hours from each of four suspected of rabies patients were collected for antemortem diagnosis of rabies. The histories of patients have been mentioned both specific and non-specific clinical symptoms such as irritability, headache, vomiting, mild fever, muscle aches, fatigue, retreat and in some cases photophobia. The main condition for entering the study was the positive results of the dFA test on their brain specimens after death that confirmed the presence of rabies virus infection in them. The saliva samples were collected from the oral cavity by a nurse on admission to the hospital via suction using a sterile syringe or a plastic dropper; while aspiration of sputum and throat is not acceptable. A healthy brain sample was also examined as the negative control. A brain sample that was positive in dFA was also considered to be the positive control.
3.3. Total RNA Extraction and Viral Complementary DNA (cDNA) Synthesis
Initially, 400 mL of each saliva samples was treated with 200 mL proteinase K and incubated for 30 minutes at 56°C; the samples were placed in a vortex every 5 to 10 minutes. Then total RNA of treated saliva and brain samples were isolated using the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The RNA was dissolved in the RNase free buffer provided with the kit. Total RNA was quantified at optical density (OD) 260 and 280 by Nanodrop 2000 spectrophotometer (Thermo Fisher, USA). A total of 50 to 100 ng viral RNA was converted into cDNA using reverse transcriptase polymerase chain reaction.
The RNA was subjected to cDNA synthesis using one µl of random hexamer (50 µM) and subjected to 70°C for five minutes and was later snap cooled on ice and briefly spun down. The RevertAid First Strand cDNA synthesis kit (Thermo scientific, Lithuania) was used for cDNA synthesis. Reverse transcriptase (Thermo scientific, Lithuania) mix was prepared and subjected to conditions 37°C for 60 minutes, 70°C for 5 minutes and chilling on ice for 5 minutes in a thermal cycler (Bio-Rad). The cDNA concentration was measured using nanodrop spectrophotometer (nanodrop technologies, CA) in ng/µL and quality was checked as a ratio of OD 260/280. The remaining parts of RNA and cDNA were frozen and stored at -70°C for further study.
3.4. PCR Amplification for Controlling of RNA Extraction Process
The quality of the RNA template was assessed by amplification of a housekeeping gene, β-actin, in each sample using PCR. Primers for the PCR assay were selected from Dacheux et al. study based on β-actin gene sequence in Genebank (accession No.: E00829) (12) and then were confirmed using the NCBI Nucleotide BLAST software (version 6.0) (Table 1).
| Name | Length, bp | TM | Sequence | Position | Product, bp | |
|---|---|---|---|---|---|---|
| β-actin-Taq1 | 25 | 64 | 73 | 5-TCACCCACACTGTGCCCATCTACGA-3 | 2206 | 295 |
| β-actin-Taq2 | 25 | 66 | 74 | 5-CAGCGGAACCGCTCATTGCCAATGG-3 | 2500 | |
Two µL cDNA was added to a 25 µL PCR mixture containing 12.5 µL 2× EasyTaq PCR SuperMix (+dye) (Yekta Tajhiz Azma, Iran) and 0.2 pM of each primer. The mixture was covered by 50 µL of light mineral oil. The initial denaturation temperature is 94°C for 3 minutes. The PCR program was 35 cycles (94°C, 30 seconds; 56°C, 45 seconds; 72°C, 40 seconds) and the final step was set at 72°C for 3 minutes using a DNA thermal cycler (Bio-Rad). The results were visualized under ultraviolet light by a transilluminator device following 1.5% agarose gel electrophoresis.
3.5. Development of Heminested RT-PCR
3.5.1. Primer Design for Heminested RT-PCR
Three primers were selected from Dacheux et al. study (12) based on conserved domains in RNA-dependent RNA polymerase (L) gene of RV strains PV (Genbank accession no. M13215) (13). The primers were confirmed using the NCBI Nucleotide BLAST software (version 6.0) (Table 2).
| Name | Length, bp | TM | Sequence | Position | Product, bp | |
|---|---|---|---|---|---|---|
| Pvo5 | 20 | 35 | 64 | 5-ATGACAGACAAYYTGAACAA-3 | 7170 | First PCR: 320, second PCR: 250 |
| Pvo8 | 22 | 40 | 69 | 5-GGTCTGATCTRTCWGARYAATA-3 | 7419 | |
| Pvo9 | 19 | 40 | 69 | 5-TGACCATTCCARCARGTNG-3 | 7489 | |
3.5.2. Heminested RT-PCR Aassay
Two µL of cDNA was used in heminested RT-PCR. The reaction mixture contained 12.5 µL of Easy TaqPCR (2U) Supermix (+dye) (10 nM NTP) + (62/5 nM MgCl2), 0.2 pM primers individually, Pvo5 and Pvo9, and 9.5 µL of PCR water to a final volume of 25 µL. The amplification was done in the thermocycler (Bio-Rad) using thermal cycling and the conditions were as follow: one cycle of polymerase activation 94°C for 3 minutes, 35 cycles with denaturation at 94°C for 30 seconds, annealing at 56°C for 45 seconds, extension at 72°C for 40 seconds, and the final extension at 72°C for 3 minutes.
Two µL of the first stage PCR product was used in the second stage. The reaction mixture contained 12.5 µL of Easy TaqPCR (2U) Supermix (+dye) (10 nM NTP) + (62/5 nM MgCl2), 0.2 pM primers individually, Pvo5 and Pvo8, and 9.5 µL of PCR water to a final volume of 25 µL. The amplification was done in the thermocycler (Bio-Rad) using thermal cycling conditions as described previously. As the negative control, no template control (NTC) reaction was also used. At the end of the amplification program, the PCR products were analyzed by 1.5% agarose gel electrophoresis with SYBR Safe stain (Invitrogen).
3.6. Development of qRT- PCR
3.6.1. Designs of Primers and Probe for qRT- PCR
The TaqMan primers and a probe were designed by Oligo Analyzer software version 3.1 and silico software (Table 3). The primers and the probe for the detection of genotype 1 of rabies virus were designed based on the Pasteur virus (PV) L protein sequence available in the GenBank database (Genbank accession No. M13215). The TaqMan probe extracted from the Dacheux et al. paper (14) was labeled at the 5’ end with the FAM as a fluorophore dye and at a 3’ end with TAMRA as a quencher molecule.
| Name Type | Length, bp | TM | nmol | Sequence | Sense | Position | Product, bp |
|---|---|---|---|---|---|---|---|
| Primer F | 26 | 63 | 25 | 5-TCTCTGTCCTAGATCAAGTGTTTGGA-3 | S | 7316 - 7341 | 98 |
| Primer R | 29 | 64 | 25 | 5-GTCTGATCTGTCTGAATAATAGACCCAGG-3 | AS | 7385 - 7413 | |
| RV5 prob (FAM/TAMRA) | 32 | 70 | - | 5-AGRGTGTTTTCYAGRACWCAYGAGTTTTTYCA-3 | S | 7384 - 7353 |
3.6.2. qRT-PCR Assay
The qRT-PCR assay was performed using the Superscript III Platinum One-Step qRT-PCR (Invitrogen) kit. The mixture contained of 5 µL 2 × reaction mix, 0.2 µL Superscript III RT/Platinum TagMix, 0.25 µL MgsO4 (50 Mm), 0.2 µL RNasin, 0.025 µL ROX, 0.15 µM RV5 probe and 0.5 µL of a 10 mM solution of sense/anti-sense primers, 0.5 µg of template cDNA added with H2O to a total of 10 µL. We used the negative control and and NTC reaction along with other samples. Thermal cycling conditions were carried out by hold one stage at 45°C for 15 minutes and hold two stages at 95°C for 3 minutes followed by 40 cycles at 95°C for 15 seconds and 61°C for 1 minute. All tests were performed in triplicate.
3.7. Evaluation of the Clinical Sensitivity and Specificity of the qRT-PCR Assay and Heminested RT-PCR Compared with FAT
Finally, we evaluated the qRT-PCR and heminested RT-PCR assays by dFA, on 50 samples obtained randomly from the repository of the National Center for Reference and Research on Rabies, Pasteur Institute of Iran. The clinical sensitivity and specificity of qRT-PCR and heminested RT-PCR methods were evaluated using the below formulae.


The qRT-PCR assay was also carried on total RNA extracted of saliva samples of four patients who had the conditions for entering the study as described above.
4. Results
4.1. Results of the Evaluation of RNA Quality Extracted Using PCR
The quality of RNA template, as a critical first step, was determined by the amplification of β-actin mRNA for each sample, with the use of β-actin-Taq1 and β-actin-Taq2 primers (Figure 1); the sequence of primers is shown in Table 2.
4.2. Sequence Analysis of Primers and Probe Used for qRT-PCR Assay
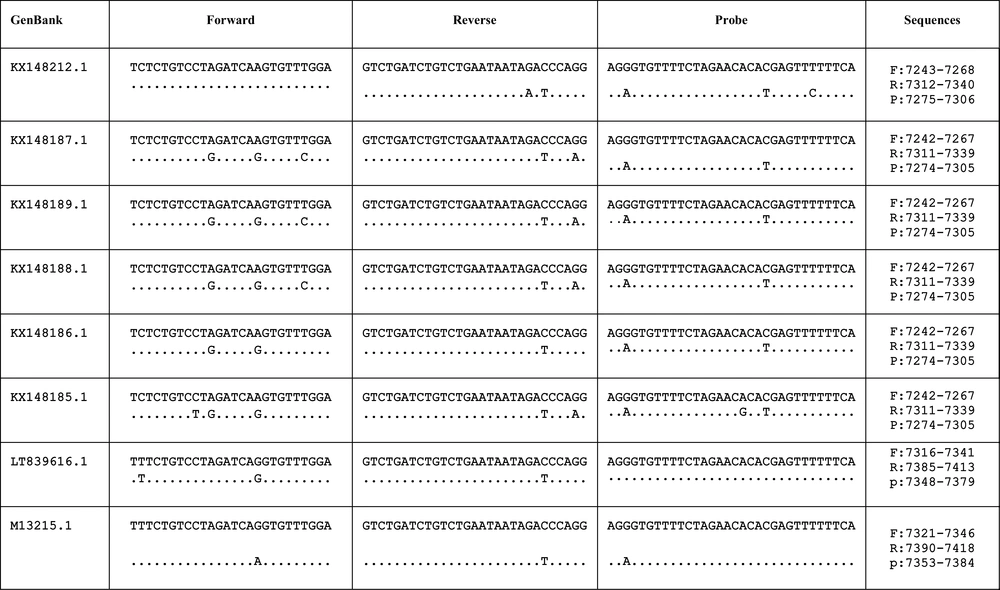
Using the NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), we confirmed a homology between the designed primers and probe based on a conserved region of the L gene sequences of the Pasteur virus fixed strain (PV) (accession number. M13215) and six RV genotypes circulating in Iran and rabies lyssavirus isolate 35009 (35009-RV-CVS, accession number. LT839616) (Figure 2).
The location and differences of designed primers based on a conserved region of the L gene sequences of six RV genotypes circulating in Iran and rabies lyssavirus isolate 35009 (35009-RV-CVS, accession number. LT839616) in comparison to the Pasteur virus fixed strain (PV) (accession number. M13215).
4.3. Quantitative Optimization of the qRT-PCR Assay and Heminested RT-PCR
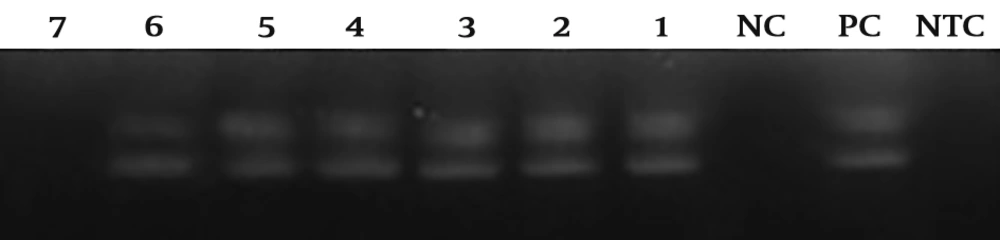
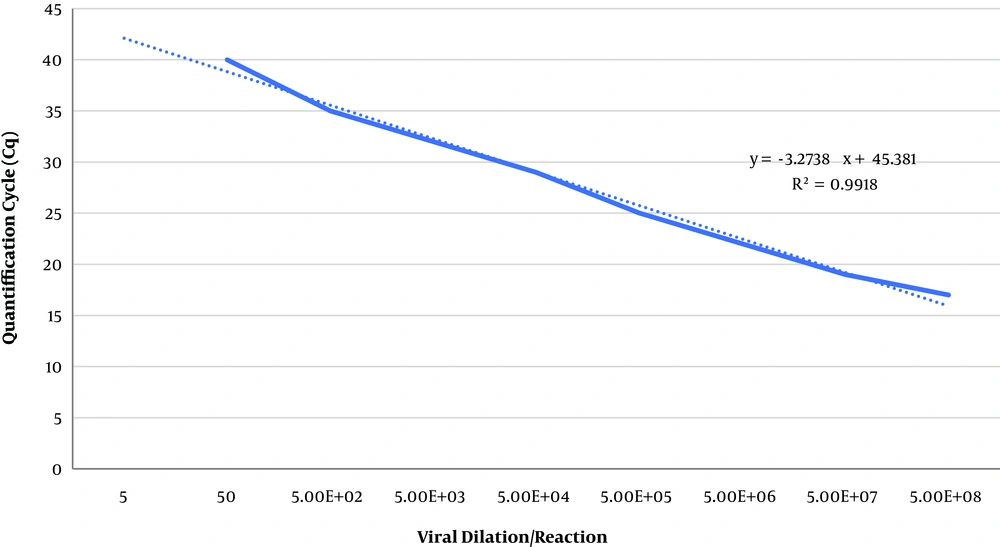
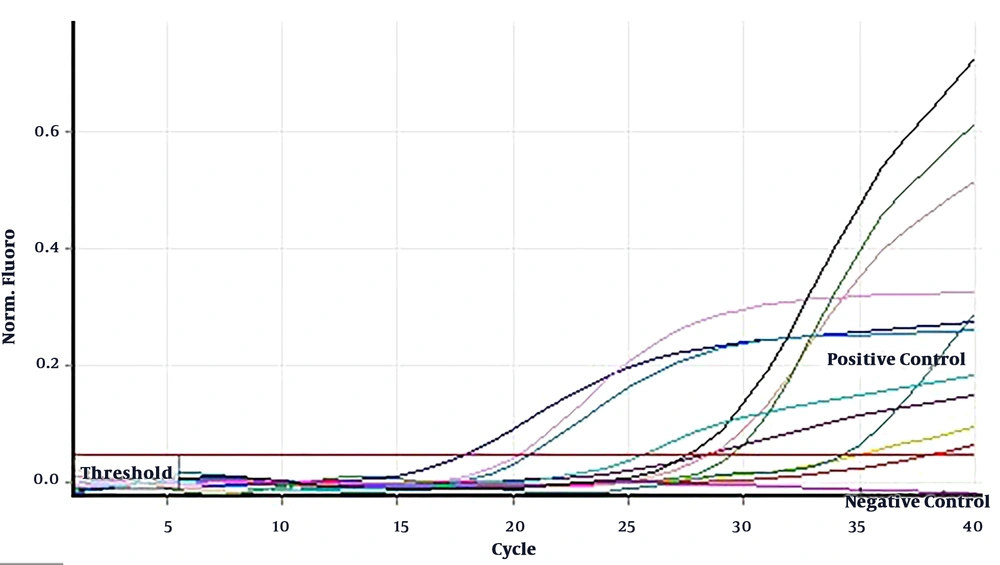
Rabies lyssavirus isolates 35009 (LD50 > 10-8) was harvested from BHK-21 cell line after about 24 hours post infection (hpi) in titer of 5 × 108 focus-forming units (FFU)/mL. Then serial dilutions of viruses were prepared up to a solution containing 50 FFU/mL. All tests were performed in duplicate. The sensitivity of qRT-PCR and heminested RT-PCR methods were about 5 × 102 and 5 × 103 FFU/mL, respectively (Figures 3 and 4).
Heminested RT-PCR analytical sensitivity results related to rabies lyssavirus isolate 35009 with 10fold dilutions from 5 × 108 to 5 × 102 FFU/mL; the test was able to identify rabies virus up to 5 × 103 FFU/mL. PC: positive control, NC: negative control with the negative sample, NTC: non-template control, 1 - 7: different viral dilution per reaction from 5 × 108 FFU/mL (well number 1) to 5 × 102 FFU/mL (well number 7). Each PCR product after each stage is clear.
Standard curves of the qRT-PCR assay related to rabies lyssavirus isolate 35009 titer. The slope equation and the correlation coefficient (R2) the linear regression curve are shown in the The The results are indicated as virus titer detected per reaction. The qRT-PCR assay was able to identify rabies virus up to 5 × 102 FFU/mL.
4.4. The Results of the Evaluation of the Clinical Sensitivity and Specificity of the qRT-PCR Assay and Heminested RT-PCR Compared with FAT
For postmortem diagnosis of rabies, the optimized heminested RT-PCR and qRT-PCR tests were highly specific (Table 3), and the sensitivity of the heminested RT-PCR and qRT-PCR were determined 94.3% and 97.14%, respectively (Figure 5 and Table 4). The qRT-PCR method has been shown more sensitive in comparison to heminested RT-PCR. On the other hand, the clinical specificity of qRT-PCR and heminested RT-PCR methods was determined 93.75% and 88.24%, respectively. For antemortem diagnosis of rabies in saliva samples submitted to the National Center for Reference and Research on Rabies, Pasteur Institute of Iran within a year, qRT-PCR showed 100% accuracy.
| Animal | Number | Results (Based on FAT and MIT) | Heminested RT-PCR | qRT-PCR | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| Brain | |||||||
| Dog | 19 | 11 | 8 | 10 | 9 | 11 | 8 |
| Cow | 13 | 11 | 2 | 10 | 3 | 10 | 3 |
| Sheep | 5 | 3 | 2 | 3 | 2 | 3 | 2 |
| Goat | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cat | 3 | 1 | 2 | 1 | 2 | 1 | 2 |
| Donkey | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| Horse | 2 | 2 | 0 | 2 | 0 | 2 | 0 |
| Wolf | 3 | 3 | 0 | 3 | 0 | 3 | 0 |
| Jackal | 2 | 2 | 0 | 2 | 0 | 2 | 0 |
| Human | 4 | 4 | 0 | 4 | 0 | 4 | 0 |
| Total | 50 | 35 | 15 | 33 | 17 | 34 | 16 |
| Saliva (Antemortem Diagnosis) | |||||||
| Human | 4 | 4 | 0 | - | - | 4 | 0 |
5. Discussion
Prominent progress has already been made in molecular biology. This allows new methods to be used for rapid and appropriate pathogen detection. For comprehensive detection of rabies virus antigen in the suspected rabid brain tissues and saliva samples in a timely manner, this study aimed to develop and validate heminested RT-PCR and qRT-PCR methods in comparison to dFA. Rapid diagnosis of rabies in suspected cases can improve the mapping of the actual prevalence of rabies in the country. In the case of antemortem diagnosis of rabies for human, it can also support the patient’s family and protect the environment and individuals in contact with the patient by the implementation of measures. The continuous development of new diagnostic methods with the aim of accurate identification of rabies and rabies-related viruses is essential to increasing information about genetic patterns, phylogeny, constant changes in the genetic pattern of the virus, spreading the different genotypes of the RV, and finally, reliable data collection (12). In remote areas where rabies is most common, it is often difficult to access the facilities, high-quality diagnostic tools, trained personnel, and unfortunately, samples are delivered to the reference lab with a delay leading to more false negatives results.
Since the clinical diagnosis of rabies is difficult and poorly reliable, especially if the history of a bite by a rabid animal is not present (15); in these cases, specialized diagnosis of rabies is required. The failure of routine diagnostic methods leads to the widespread use of molecular methods for detecting rabies. Molecular techniques are based on genome replication and identification of the viral nucleic acids (16). The molecular analysis techniques, because of their high sensitivity and repeatability, have the ability to be used instead of FAT as an invasive test or other time-consuming methods such as mouse inoculation test (MIT) (12). The MIT is performed on a number of live animals that are not in molecular methods. Also, the sensitivity of dFA is reduced during the use of decomposed brain samples or in low viral load states (17). The endemic areas in terms of rabies have usually warm-dry climate. The probability of decomposition and degradation of the brain tissue is increased in this type of climate. The studies have shown that dFA on decomposed carcasses demonstrates false-negative results after four days of incubation at 25°C, while viral RNA can be detected after 37 days at 36°C (18). Thus methods using PCR technology such as qRT-PCR and heminested RT-PCR are reliable, sensitive, and rapid tools that can be used to solve this problem.
heminested RT-PCR has been described to the diagnosis of rabies for over a decade. The final detection time can be shortened by using the heminested RT-PCR method. Predominantly, all studies have increased the sensitivity and specificity of molecular diagnostic techniques by targeting the N gene as the most conserved gene of rabies virus to solve various PCR problems. Recently, the mean detection range of a wide range of samples collected from human, livestock, and wildlife, was improved by the detection of the longest virus gene (L polymerase gene) (12). In this study, the effective factors in the reaction were optimized such as time, pattern, size, and reaction temperature. Echevarria et al. in the study conducted on the bat populations detected viral RNA even in healthy bats. Of 33 brains tissues of captive bats, only five cases have been reported as positive results by routine diagnostic methods, while a molecular-based method for genome detection found 15 rabies-positive cases (19). Therefore, these methods can be used as reliable epidemiological tools for screening in suspicious populations.
In the study, the clinical sensitivity and specificity of the heminested RT-PCR method compared with dFA and MIT on 50 brain tissue samples were determined at 94.3% and 88.24%, respectively. Internal control was used for validation of the RNA extraction process and avoid any false negative findings. The heminested RT-PCR method as the reliable and alternative technique can be used in laboratory diagnosis of rabies infection when other methods are not feasible. This method does not require dedicated devices and it is capable of running with a simple protocol and a thermocycler. Furthermore, this method usually provides highly sensitive and specific results, which can be confirmed by the dFA (17). Despite the fact that real time PCR needs a specific device and the high cost of dedicated probe design, this method has certain advantages over Himenested RT-PCR, such as sensitivity, large detection range, and low risk of infection, as well as eliminating all inherent defects (20). In this study, the clinical sensitivity and specificity of the qRT-PCR method compared with dFA and MIT on 50 brain tissue samples were determined 97.14% and 93.75%, respectively, which are higher than heminested RT-PCR launched. Another disadvantage of heminested RT-PCR is the analyzing of the amplification products in the agarose gel electrophoresis followed by observing the results under UV lamp and sometimes even in the presence of a carcinogen such as ethidium bromide (20).
This study was consistent with the results of the other studies, who have shown that qRT-PCR is the accurate molecular diagnosis method in terms of technical features, quantitative and qualitative detection capabilities, and simultaneous reproduction (21). Certainly, the selection of probes and primers is the most important issue that should be considered in development of qRT-PCR to obtain reliable results. However, due to the high genetic variation of RV, development of a single sensitive qRT-PCR assay covering all widely known phylogroups remains a challenge (22). The primer and probe set of this study were selected by reviewing the articles and according to the range of detection, target sequence, the number and type of degenerate bases in the sequence, the number of their mismatches after BLASTing, and the location of these heterogeneities (the significance of the sequence at the end of 3’). In the current study, we developed qRT-PCR by amplifying an area shorter than target sequence in the genome; this high accuracy is useful for detection of the low virus titers even before the patient’s antibodies reach to a detectable level in the intravitam diagnosis of rabies.
5.1. Conclusions
As a result of the study, heminested RT-PCR and qRT-PCR not only can be used with a high confidence level for diagnosis but also it is an opportunity for sample screening and pathogenicity studies, epidemiology, and differentiation of lyssavirus genotypes. The set of the probe and primers designed for qRT-PCR has the ability to identify a wide range of positive samples due to the shortage of product length, high specificity and sensitivity, deletion of degenerate and error from sequences. The results of tests carried out on samples with different type and geographically position in this study approved this method as a practical alternative for classical methods such as dFA and MIT.