1. Background
Acinetobacter baumannii is considered one of the highly troublesome pathogens to health services around the world due to immense ability to cause endemic and epidemic nosocomial infections, especially in hospital intensive care units (1, 2). This bacterium causes pneumonia associated with ventilator, bloodstream infections, secondary meningitis, wound infections, soft tissue and urinary tract (3, 4). According to the study from “Center for Disease Prevention and Treatment in Europe”, A. baumannii is responsible for 8.7% of all cases of pneumonia and lower respiratory tract infections and 1.4% of all blood infections (5) with the 44% and 72% mortality rate to meningitis and pneumonia in some reports, respectively (6, 7). The majority of the species populations are currently resistant to carbapenem (8) and also increased resistance to colistin and tigecycline have been observed (9). Moreover, these strains are incurable due to pan-drug resistant (PDR) to all FDA-approved antibiotics (10).
Vaccination of at-risk patients is a promising strategy to reduce the rate of infections in the absence of effective antibiotics against A. baumannii. Protection against A. baumannii infectious bacteria is currently achieved through active or inactivated immunization of rats with antigens such as OmpA (11), Ata (12), and Bap (13), which indicate an important and accessible vaccination role. However, the dissolution, variety, and abundance of these antigens are significant barriers for providing a widely available protective vaccine. Therefore, new studies are needed to design and produce suitable antigens as candidates for a vaccine in order to evaluate their antigenic and immunological properties. Although whole cell-based vaccines can stimulate antibodies to several bacterial outer membrane proteins, they cannot be useful due to the presence of several bacterial components (14). Therefore, the use of recombinant chimeric proteins harboring antigenic compartment could be considered a suitable remedy for the protection against the lethal bacteria, especially A. baumannii.
Moreover, A. baumannii outer membrane protein A (AbOmpA) is one of the important virulence factors for its pathogenesis, which is highly conserved across a wide variety of species of Gram-negative bacteria. The AbOmpA protein has been shown to play a critical role in a number of interactions, including adherence to epithelial cells, induction of apoptosis in host cells as well as differentiation of host immune cells (15-18).
2. Objectives
In the overall point of view, the purpose of the present study is to provide recombinant OmpA fragment containing superficial epitopes of B-cell stimulant amino acids from 240 to 356 residues in order to investigate the immunogenicity of the desired protein.
3. Methods
3.1. Genomic DNA Isolation, Gene Amplification and Cloning
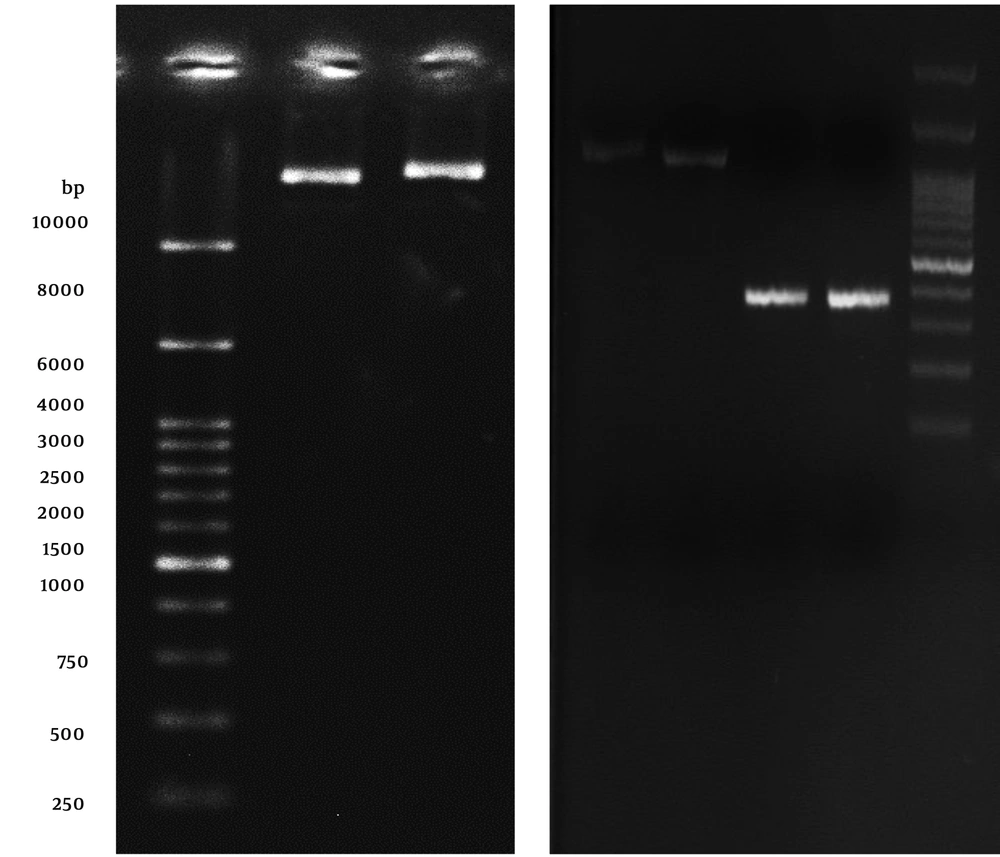
The A. baumannii ATCC 19606 was kindly gifted from microbiology department, Shahid Beheshti University of Medical Sciences. To confirm the nature of the isolates, several tests including Gram’s staining, motility, oxidase activity, TSI, and ability to grow in 42°C was performed, according to the standard protocols. The total genomic DNA was extracted via total genome extraction kit (Geneall-Korea). Then the isolated DNA was dissolved in 200 µL TE (Tris 10mM, EDTA, 1mM) buffer for further analysis. The 240 - 356 fragment of OmpA gene sequence was amplified via PCR technique using primers containing NcoI and HindIII restriction sites (indicated in bold): (F: 5’-CATCCATGGTAACCCATGGAACATC-3’ and 5’-TTCAAGCTTCAGCAGCTCAAAAGC-3’) and the following procedure: initial denaturation at 94°C for 5 minutes, followed by 30 cycles of 94°C for 45 seconds, 56°C for 45 seconds, and 72°C for 60 seconds; and a final extension at 72°C for 8 minutes. The PCR products were purified, digested with NcoI and HindIII, and then isolated on 1% agarose gel.
The double-digested PCR products were ligated to NcoI/HindIII-digested pET-28a (+) (Novagen, Darmstadt, Germany), and the ligation mixture was transformed into the pre-cultured cloning host, Escherichia coli strain DH5-alpha. Chemical heat shock DNA transformation was performed for 240 - 356 ligated fragments. Finally, plasmid DNA was purified using the Bio-basic plasmid purification kits (Canada). To confirm the cloning procedure, the recombinant plasmid was isolated and restriction site digestion was performed.
3.2. OmpA 240 - 356 Fragment Expression and Purification
Expression of OmpA 240 - 356 fragment was performed in E. coli BL21 (DE3). The E. coli cells were grown aerobically in 200 RPM shaking condition at 37°C in LB medium containing Kanamycin for an overnight. Then after subculturing in the fresh medium, until the optical density (OD at 600 nm) reached 0.6 nm, the cells were inducted for protein expression using 1mM IPTG for 4 hours. Then the cells were harvested by centrifugation and re-suspended in a small volume of lysis buffer containing 150 mM sodium chloride, 1.0% Triton X-100, and 50 mM Tris pH 8.0. Cell disruption was performed via ultrasound method for 5 × 30 seconds on ice with 30-second intervals. Finally, the cell debris was removed by centrifugation (12000 × g, 15 minutes, 4°C) and the supernatant was kept at 4°C for further analysis.
The bacterial supernatant was loaded on to the Ni/NTA affinity column chromatography (Takara-Japan) and eluted with 250 mM imidazole. Before loading, the column was equilibrated with the lysis buffer containing 10 mM imidazole. The cell supernatant was loaded to the column. Subsequently, the column was washed with buffers with increasing concentrations of imidazole (20, 50, 100, and 250 mM) to elute the purified proteins.
The protein concentration was determined by Bradford assay, according to the manufacturer’s protocol. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was finally performed, according to the standard protocols. Also, the expression was further confirmed via Western Blotting using 6x-His Tag Antibody, according to the general procedure. In this propose, 15 micrograms of each sample were resolved on a 12% acrylamide gel and either visualized using Coomassie staining or transferred to nitrocellulose for Western blotting with a 1:1000 dilution of murine anti-6x-His antibody (Sigma # H1029). The double-digested PCR products were ligated to NcoI/HindIII-digested pET-28a (+) (Novagen, Darmstadt, Germany), and the ligation mixture was transformed into the pre-cultured cloning host, E. coli strain DH5-alpha. Chemical heat shock DNA transformation was performed for 240 - 356 ligated fragments. Finally, plasmid DNA was purified using the Bio-basic plasmid purification kits (Canada). To confirm the cloning procedure, the recombinant plasmid was isolated and restriction site digestion was performed.
4. Results
4.1. AbOmpA240 - 356 Cloning in E. coli
A 348-bp fragment encoding a polypeptide containing 116 amino acids from an end part of A. baumannii OmpA gene (AbOmpA240-356) was successfully amplified and cloned into the pET-28a (+) vector at the NcoI and HindIII restriction sites (Figure 1), which incorporates a 6His-tag onto the C-termini of the protein fragment. The molecular weight of AbOmpA240-356 was estimated to be 17425.74 Da, and the theoretical pI of the protein fragment was calculated to be 9.22 via ExPASy (http://www.expasy.org/cgi-bin/protparam). Furthermore, the estimated half-life for the protein was predicted to be 30 hours in mammalian reticulocytes (in vitro), > 20 hours in yeast (in vivo) and > 10 hours in E. coli (in vivo) environment. The instability index for the recombinant the OmpA240-356 was estimated as 28.20. The extinction coefficient value at 280 nm, aliphatic index, and grand average hydropathy (GRAVY) for the protein was predicted as 11460 M-1cm-1, 82.67, and -0.447, respectively.
4.2. Expression and Purification of the Recombinant OmpA240-356 in E. coli
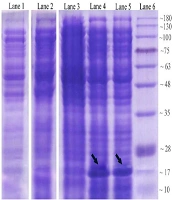
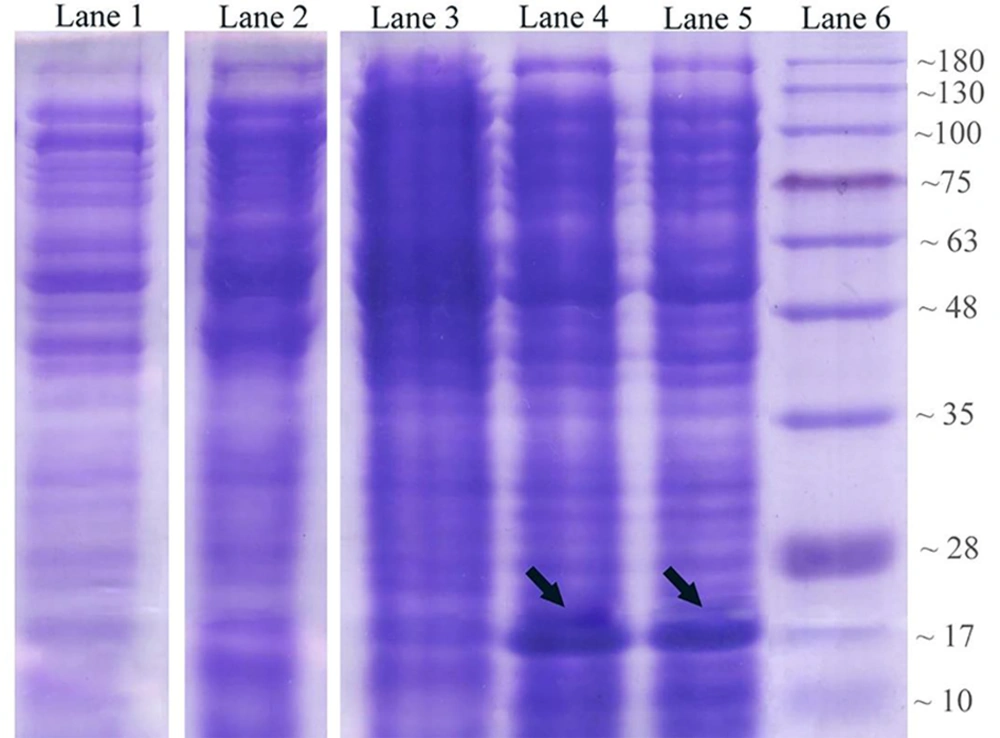
The expression process of recombinant OmpA240-356 in E. coli as the host cell was assessed. Coomassie-stained gels of the soluble fractions of IPTG un-induced and induced bacterial culture in the presence of Kanamycin demonstrated that a band of approximately 17 kDa, which is consistent with a 6His-tagged form of AbOmpA240-356, is detectable (Figure 2A). Levels of the induced protein in the soluble fraction appeared to peak approximately 6 hours post-induction.
Analysis of OmpA240-356 fragment protein expression at different harvesting time points in 2.5 mmol/L IPTG concentration. Lane 1: control negative Strain (OmpA--); lane 2: un-induced host cell supernatant; lane 3: two hours after the induction; lane 4: six hours after the induction; lane 5: eight hours after induction via 2.5 mM IPTG; lane 6: protein marker.
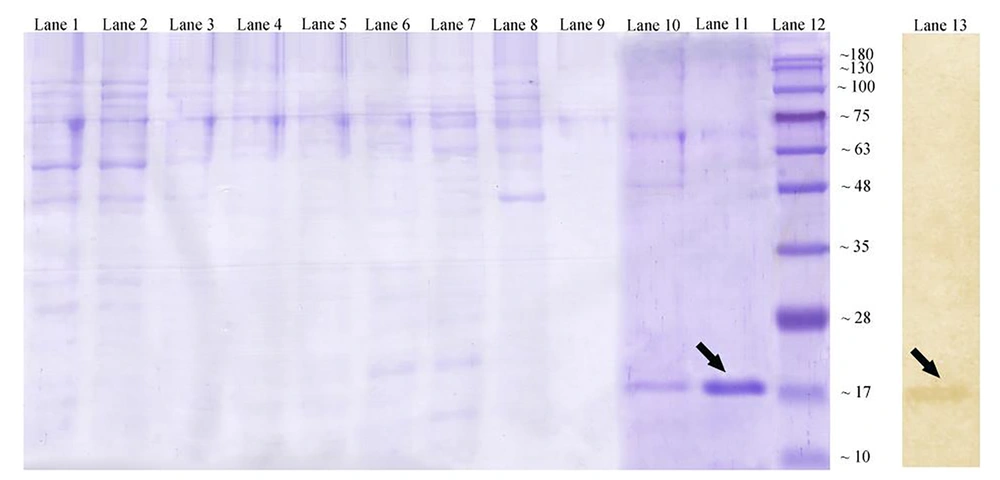
To gain the purified AbOmpA240-356, cell lysates were collected at 6 hours post-induction and applied to a nickel matrix for affinity purification. As mentioned in Figure 3, AbOmpA240-356 protein fraction is successfully purified in 250 mM imidazole. After elution from the nickel matrix, AbOmpA240-356 was highly pure (Figure 3, lane 11). Since SDS-PAGE method only shows protein profiles of the cell with the approximate molecular weight, to ensure the accuracy of the nature of the protein, the western blotting method was used. So, to verify that the induced protein was the 6His-tagged product encoded by the plasmid, replicate gels containing soluble fractions were assessed with an anti-6His-tag antibody. As shown in Figure 3A, the antibody recognized the induced protein of approximately 17 kDa in the soluble fraction.
Affinity chromatography for OmpA240-356. Coomassie-stained gel showing lane 1: washing fractions buffer containing 20 mmol/L imidazole; lane 2: washing fractions buffer containing 50 mmol/L imidazole; lanes 6 - 9: washing fractions buffer containing 100 mmol/L imidazole; lane 10: washing fractions buffer containing 250 mmol/L imidazole; lane 11: eluted recombinant protein with standard method. Lane 12: protein marker (Abcam (10 - 180 kDa) # ab116027-3); lane 13: western blotting technique of the expressed recombinant OmpA240-356 protein by using mouse monoclonal anti-His-Tag antibody.
5. Discussion
Acinetobacter baumannii is a fast dispreading nosocomial pathogen that has gained attention in the last two decades worldwide (18, 19) due to its multidrug resistance nature (1, 3, 19, 20). Although colistin is currently used to overcome the pathogen-induced infection, there are several reports of the emergence of A. baumannii colistin-resistant strains (9, 21, 22). Hence, other than antibiotics, pre-hospitalizing vaccination of patients in intensive care units appears as a promising and effective treatment option to overcome this issue.
Nowadays, producing of putative conformational antigen via recombinant protein technology to stimulate immunity instead of the full-sized protein would be helpful to reduce the side effects for vaccination targeting designs. In this case, shortening and more specificity of the desired piece will facilitate the work and possibly increase the protection. Given the fact, protective immunity toward A. baumannii is mainly dependent on antibody production, thereby introducing immune-stimulating B-cell epitopes may be helpful.
Previous studies have shown that OmpA, Omp34kDa, OprC, OprB, OXA-23, and siderophore protein are the most powerful immunogenic protein against A. baumannii infection (11, 23). Reports showed that vaccination of mice with recombinant OmpA as a major foreign membrane antigen promotes increased resistance of the mice to infection, reduces the number of bacteria, and produces hemorrhagic immune responses. The researchers have suggested that OmpA can be a promising candidate for the prevention and control of infection caused by antibiotic-resistant A. baumannii (9). In another study, the AbOmpA was cloned in E. coli and purified via affinity chromatography, which was capable of inducing cell death in the A549 cell line and producing a polyclonal antibody against OmpA (24). Moreover, it has been shown that AbOmpA is mainly involved in the multi-drug resistance of the bacterium (25). The immunogenic epitopes in AbOmpA were first introduced by Lynn and his colleagues. According to the information presented in this study, a piece of OmpA that contains superficial epitopes of B-cell stimulus at the interval between nucleotides 718 - 1071 (26). In the present study, we describe the overexpression and purification of stable AbOmpA240-356 in E. coli as a host cell.
5.1. Conclusions
This report describes a method for expression and purification of stable AbOmpA240-356 from soluble supernatant. Consequently, this work may facilitate further characterization of AbOmpA240-356 for the development of novel antibacterial therapies, such as antibacterial peptides for targeting this protein or even using it as a successful vaccine.