1. Background
Ventilator-associated pneumonia (VAP), pneumonia occurred 48 - 72 hours after intubation and mechanical ventilation, is the common infectious disease related to the intensive care unit (ICU) (1). VAP occurs in 9% - 27% of mechanically ventilated patients in the ICU and is characterized by the presence of a new or progressive infiltrate, fever, altered white blood cell, and changes in sputum properties (2). The development of VAP is mainly determined by the complex interplay between endotracheal tubing, the presence and virulence intensity of resident or invading bacteria, and host immunity (3). The type of VAP-causing organism is associated with duration of mechanical ventilation. Early VAP is usually caused by Streptococcus pneumoniae (Pneumococcus), Haemophilus influenzae, and meticillin-sensitive Staphylococcus aureus (MSSA), whereas late onset of VAP is caused by multi-drug resistant Acinetobacter, Pseudomonas aeruginosa, Klebsiella, and MRSA (4). Pneumococcus is considerate as one of the most prevalent nasopharynx normal flora with a potential to cause invasive pneumococcal disease (IPD) (5, 6).
As a common colonizer of human upper respiratory tract and also the most common cause of community acquired pneumonia (CAP), Pneumococcus, is frequently isolated in early onset VAP (2, 7). There is no gold standard method for the precise diagnosis of VAP yet (8), and accordingly, the precise diagnosis of Pneumococcus from VAP is difficult due to the microbial normal flora of nasopharynx (9). There is no report regarding the prevalence of Pneumococcus in VAP in Iran. Regarding the potential of real-time PCR for specific identification of S. pneumoniae (9, 10), the aim of the present study was to determine the prevalence of bacterial agents of VAP, and to evaluate the presence of S. pneumoniae in VAP- confirmed ICU patients by real-time PCR.
2. Methods
2.1. Ethics Statement
As clinical specimens were obtained routinely during diagnosis and treatment procedure, there was no need for any particular ethics consideration; in addition, this study was approved by the Ethics Committee of Damghan Azad University Bioethics Committee.2.2. Study Setting and Subjects
In this cross-sectional study, a total of 90 ICU admitted patients suspected of pneumonia were studied during March 2016 to March 2017 in Tehran, Iran. Among these, 60 patients were selected based on the following VAP criteria.
Inclusion criteria were a positive recent chest X-ray radiograph as well as the clinical and laboratory findings including fever, cough, new pulmonary infiltration, increase body temperature to 38.3°C or higher. Any antibiotics therapy before sampling was considered as the exclusion criteria and based on this limitation, 10 patients were excluded and 50 participants were included in the study. The patient’s age, gender, and other demographic data were examined.
Prior to antibiotic therapy, tracheal aspirates were obtained from patients and transported to the research laboratory in Baqiyatallah University of Medical Sciences for further analyses.
2.3. Direct Microscopy Examination
Tracheal aspirate smears were prepared and stained with Gram staining. Samples with more than 10 epithelial cells/lpf were discarded. Finally, 10 samples were excluded and 50 samples were selected for WBC count and future analysis.
2.4. Biochemical Identification
According to the routine standard protocol, primary bacterial identification was done by biochemical tests after culturing the specimens on sheep blood agar, chocolate agar, and MacConkey agar and incubated for 24 hours at 37°C (11).
2.5. DNA Extraction
To evaluate the presence of pneumococcus in samples, real-time PCR was performed. At first, genomic DNA was extracted from specimens using the Mini Kit (Qiagen, Hilden, Germany), according to manufacturer’s instruction. The S. pneumoniae ATCC 49619 was used as standard control. The optical density (OD) of extracted DNA was determined at 260 nanometer by a NanoDrop 1000 (Thermo Scientific, USA).
2.6. Real-time PCR
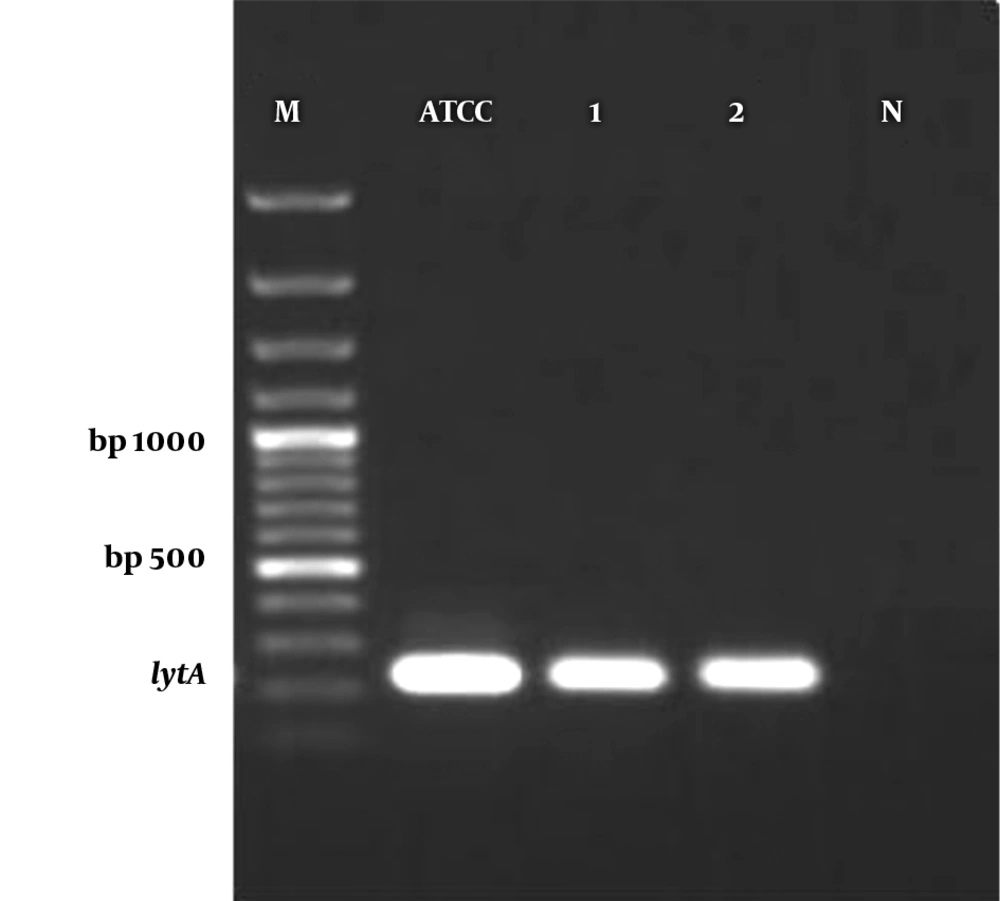
Real-time PCR assay was carried out using TaqMan universal PCR master mix and primers for lytA gene (the autolysin gene). The primers sequences (BioNEER, Korea) are (5'-3') f-ACGCAATCTAGCAGATGAAGCA-r and f-TCGTGCGTTTTAATTCCAGCT-r and the probe sequence is 5'-FAM-GCCGAAAACGCTTGATACAGGGAG-3'-BHQ1 (9). The specificity of primers and probe sequences was determined by comparison of available sequences, using the BLAST database search program (http://www.ncbi.nlm.nih.gov/BLAST). The reactions were conducted in a ABI 7500 real-time PCR machine (Applied Biosystems, USA). Firstly, the standard curve for lytA gene was assessed with a tenfold serial dilution of S. pneumoniae ATCC 49619. According to the standard curve and y-intercept, samples which did not display the fluorescent signal earlier than the Ct of 37 were considered as negative. The efficacy of the real-time PCR was calculated by the following formula: E = 10(−1/slop) - 1 (12). After the optimization and qualification of standards curves, the main reaction was performed according to the following procedure. All assays were performed in a total volume of 25 µL consisting of 12.5 μL of 2 × TaqMan universal master mix (Applied Biosystems, Foster City, CA), 0.1 mM probe, 400 nM primers, and 2 ng DNA in distilled RNase/DNase-free water. The PCR condition was as follows: holding at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 20 s, followed by annealing at 52°C for 40 s, and extension at 72°C for 20 s. A negative control was included in each run. The specificity of the real-time PCR was checked by gel electrophoresis for products as well as the post-PCR melting-curve analysis performed under the following conditions: temperature starting at 60°C for 10s followed by 0.5°C/10 s rising up to 95°C.
2.7. Detection Limit Assessment of the Real-time PCR
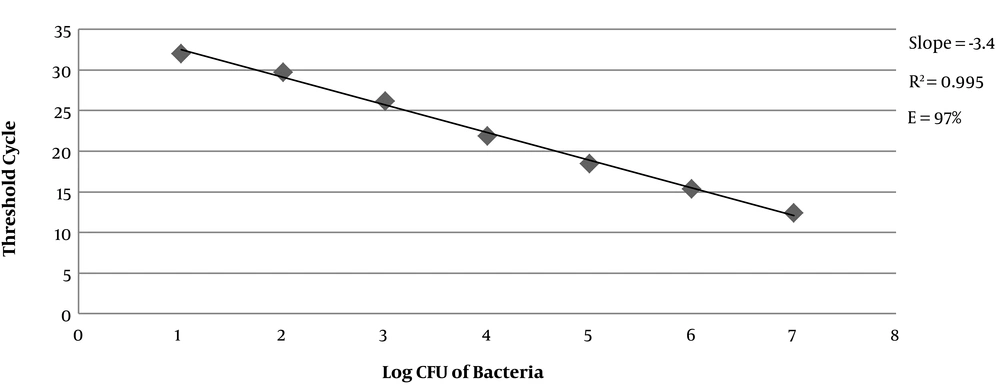
To determine the detection range of real-time PCR, a standard curve for S. pneumoniae ATCC 49619 was generated as follows: S. pneumoniae was grown aerobically in TSB medium at 37°C for 4 hours to reach the logarithmic phase. The culture was diluted with physiological saline (pH = 7) until it reached a 0.5 McFarland standard, representing approximately 108 CFU/mL. Starting from this concentration, 10-fold serial dilutions were prepared in the physiological saline, and the number of CFU was determined by inoculate 100 μL of each dilution onto sheep blood agar plates with the overnight aerobic incubation at 37°C. One milliliter of each dilution (101 - 107 CFU/mL) was used for DNA extraction, followed by amplification as described above. The calculated CT values were then plotted against the numbers of bacteria.
3. Results
3.1. Demographic Data
During the 12-month study, 50 cases of VAP (35 (70%) men and 15 (30%) women) with a range of 50 to 90 years were studied. Table 1 shows the distribution of age and gender of patients. As shown in Table 1, VAP patients were dominantly placed in the 70 - 80 years old age group (42%).
| Age Group, y | Male | Female | Total (%) |
|---|---|---|---|
| 50 - 60 | 3 | 0 | 3 (6) |
| 60 - 70 | 11 | 5 | 16 (32) |
| 70 - 80 | 14 | 7 | 21 (42) |
| 80 - 90 | 7 | 3 | 10 (20) |
| Total | 35 | 15 | 50 (100) |
3.2. Direct Microscopy and Microbial Patterns
All samples had more than 10 WBC/lpf in microscopy examination, indicating an inflammation/infection. According to the culture, 40 out of 50 samples (80%) resulted in a pure isolate, in which Klebsiella pneumoniae and Pseudomonas spp. were the most and least prevalent bacteria, respectively. The culture results for bacterial isolation are shown in Table 2. In addition, in seven samples, Candida spp. was isolated as a pure culture and all cultures were negative for S. pneumoniae.
| Type of Bacteria | No. of Isolates (%) |
|---|---|
| Klebsiellapneumoniae | 13 (26) |
| Acinetobacter ssp. | 9 (18) |
| Escherichia coli | 7 (14) |
| Coagulase negative Staphylococci ssp. | 5 (10) |
| Streptococcus ssp. | 3 (6) |
| S. aureus | 2 (4) |
| Pseudomonas ssp. | 1 (2) |
| No growth | 3 (6) |
3.3. S. pneumoniae Identification by Real-time PCR
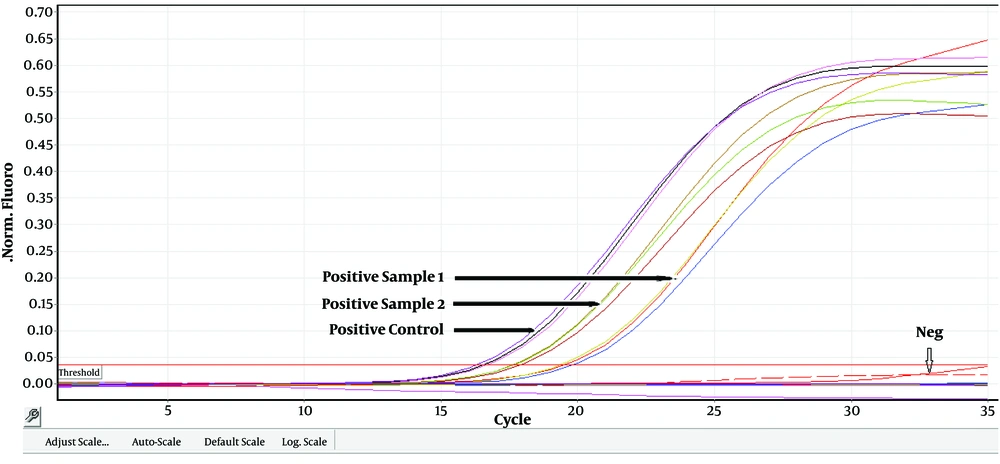
According to the standard curve for the positive control, 101, 102, 103, 104, 105, 106 and 107 CFU/reaction concentrations provided CT values of 31.96 ± 0.2, 29.69 ± 0.2, 26.17 ± 0.3, 21.87 ± 0.3, 18.44 ± 0.3, 15.34 ± 0.4, and 12.35 ± 0.2, respectively. The efficacy of the real-time PCR was between 95% to 100% (Figure 1). Furthermore, all standard dilutions had one band in the gel electrophoresis (Figure 2). In addition, samples without a fluorescent signal before 37 cycles were considered negative. In two samples (4%), S. pneumoniae were identified by real-time PCR method. Based on CT values obtained for these two positive samples, the rate of S. pneumoniae was calculated 4 × 104 and 1.6 × 105 CFU/mL (Figure 3).
4. Discussion
An accurate, in-time diagnosis of pneumococcal VAP, as an early-onset VAP agent, has been frequently hampered not only by the difficulties in bacterial isolation from the patient, but also by the misidentification of pneumococcus-like Viridans streptococci as S. pneumoniae, especially in isolating the pathogen from the respiratory tract (13-15). To address this issue, researchers have introduced molecular methods including real-time PCR (16). This study tried to determine the prevalence of bacterial agents of VAP, and to evaluate the presence of S. pneumoniae in VAP- confirmed ICU patients by real-time PCR. The results showed that Klebsiella pneumoniae followed by Acinetobacter ssp. are the most prevalent VAP bacterial agents. All samples were negative for S. pneumoniae in culture; however, in real-time PCR, two samples (4%) were positive for this pathogen with 4 × 104 and 1.6 × 105 CFU/mL bacterial load. These two samples contained 10 and 12 WBC/lpf, respectively. Age is one of the important risk factors for VAP. Similar to other studies, our results showed that VAP is more prevalent in elderly patients and is more seen in men than women (17). To date, several S. pneumoniae genes have been used to detect the pathogen, among which three most applied pneumococcal genes were lytA, ply, and psaA that encode autolysin, pneumolysin, and surface adhesion A, respectively (18). In a study performed by Adams et al., the specificity levels of lytA, psaA, and ply for detection of S. pneumoniae were reported 100%, 98%, and 81%, respectively (12). According to the high sensitivity and specificity of lytA, we selected this gene to investigate the presence and quantification of S. pneumoniae in respiratory specimens of VAP patients. In this study, based on culture, K. pneumoniae and Acinetobacter ssp. were the most common pathogens isolated from VAP patients. In the Chi et al., study, S. aureus and A. baumanii were the first and second causative pathogens of VAP (13). According to the SENTRY antimicrobial surveillance program operated in the US, Europe, and South America, P. aeruginosa (27%) was reported as the most common isolated VAP pathogen, followed by S. aureus (20%) and Acinetobacter ssp. (14%) (19).
In general, there are few studies in Iran evaluating the frequency of bacterial pathogens in VAP patients. A study on the bacterial prevalence in VAP patients in Iran, Enterobacteriaceae (35.4%), S. aureus (20.7%), and Staphylococci spp. (14.7%), P. aeruginosa (11.3%), A. baumannii (9.4%) and Corynebacterium spp. (7.5%) were the most prevalent (16). In another study in Iran, the most common isolated organisms were Klebsiella spp. (36.36%), Pseudomonas spp. (27.27), Acinetobacter spp. (27.27), and E. coli (9.09%) (20). The results of this study are consistent with our findings. In addition, A. baumannii, Methicillin-resistant S. aureus, and P. aeruginosa were reported as the most prevalent bacteria isolated from VAP in ICU patents (21). Beside the patient’s characteristics, the time of VAP development may also determine the causative pathogen. According to reports, S. pneumoniae is responsible for relatively low rates (4.1%) of VAP (early VAP) worldwide, for which smoking, chronic obstructive pulmonary disease (COPD), and the absence of prior antibiotic therapy were the main risk factors (2, 4, 22).
The results of the real-time PCR showed that, although two samples were reported to be negative in culture, they were positive with higher counts than the threshold (104 CFU/mL) (2). Negative culture results of S. pneumoniae could be due to the fragile nature of the organism, previous antibiotic therapy of patients, low experienced technician, inaccurate laboratory handling, absence of bile solubility test, the presence of optochin-resistance strains, and more importantly, low efficacy of optochin disks (misidentification with non-pathogenic respiratory normal bacteria such as Viridans Streptococci group) (23). In addition, the presence of many respiratory pathogens, including pneumococci as a part of normal respiratory flora in one hand, and the non-quantitative nature of conventional PCR and culture methods on the other hand, have made the issue more complicated. Accordingly, the accurate detection of respiratory infections is still a critical diagnostic problem, and therefore, a method being able to detect and quantify simultaneously the causative pathogen is appreciable (22, 24, 25). To overcome these limitations, due to the ability of diagnostic species-specific real-time PCR, the present study used a quantitative real-time PCR for detecting and quantifying of Pneumococcal VAP with advantages of saving time and analyzing directly on clinical VAP specimen. Our study has some limitations, including the low numbers of clinical samples, possibility of contamination of tracheal aspirates with upper respiratory tract normal flora, and lack of full patient information.
4.1. Conclusion
Although a few studies have investigated the microbial agents of involved VAP in Iran, none have determined the rate of pneumococcal VAP by real-time PCR; thus, this is the first report of pneumococcal VAP in ICU patients in Iran. Our results showed that to detect pneumococcal VAP the real-time PCR has a higher sensitivity than microbial culture. Accordingly, more studies for optimizing cultural methods and commercialize diagnostic real-time PCR for pneumococcal VAP detection could be appreciated.