1. Background
S. aureus is a Gram-positive, non-motile, facultatively anaerobic, and ubiquitous coccus found predominantly on the skin and mucous membranes (1). This bacterium is recognized as one of the most important causes of hospital-acquired infections. S. aureus infections, often in acute and pyogenic forms, are highly prevalent and they are also capable of spreading to other tissues of the body through bacteremia (2, 3). The spectrum of diseases caused by this bacterium includes various skin infections such as folliculitis, acne, abscess, and wound infection, as well as life-threatening diseases such as sepsis, toxic shock syndrome (TSS), osteomyelitis, endocarditis, bacteremia, and food poisoning (4). In this respect, antibiotic resistance is still a major concern in the treatment of infectious diseases due to the misuse of conventional antibiotics (5). These bacteria turn to MRSA strains with the acquisition of the mecA gene (6). The mobile genetic elements (MGEs) including plasmids, integrons, and transposons are also responsible for the acquisition and further spread of drug resistance genes (7). The role of integrons, as MGEs, has been recognized as a mobile genetic mechanism in the horizontal transfer of antibiotic resistance genes. Thus, the integrons are of importance due to the presence of a particular recombinant system leading to the introduction and expression of various genetic cassettes. Moreover, the horizontal transfer of the integrons is considered to be the most effective method of spreading antibiotic resistance genes and producing multidrug-resistant (MDRs) isolates (8-10). Besides, various classes of integrons have been identified based on the difference in their integrase genes. Class I integrons have the highest frequency among Gram-positive and Gram-negative bacteria isolated from clinical specimens. Class II integrons have a lower prevalence rate than class I integrons and they are more reported in Gram-negative bacteria. Other classes of integrons have a much lower incidence (11, 12). Structurally, the integrons have conserved 5’ and 3’ ends, an integrase gene, and a variable central domain between these regions wherein the gene cassettes are located (13). The integrons also carry resistance genes in the gene cassette via integration within plasmids, chromosomes, and transposons. Abundant antibiotic resistance genes are further transferred by the integrons and these genes play a role in generating resistance to a wide spectrum of antibiotics, including aminoglycosides, β-lactams, macrolides, sulfonamides, and chloramphenicol (14).
2. Objectives
We found no recent comprehensive study in the city of Kermanshah, Iran, concerning the frequency of class I and II integrons in S. aureus isolates. Thus, the main purpose of this study was to determine the frequency of class I and II integrons in methicillin-resistant S. aureus (MRSA) and methicillin-sensitive S. aureus (MSSA) isolates from clinical specimens in Kermanshah, Iran.
3. Methods
3.1. Bacterial Isolates and Identification
The present descriptive cross-sectional study was conducted on 86 strains of S. aureus isolated from different clinical specimens (including blood, wound, urine, trachea, catheter, synovial fluid, and abscess) at Imam Reza Hospital in the city of Kermanshah, Iran, from September 2017 to July 2018. The Ethics Committee of the Kermanshah University of Medical Sciences approved the study protocol (KUMS.REC.1395.515). The isolates were identified using standard biochemical tests including Gram staining, oxidase, catalase, DNAase (the Merck Group, Germany), coagulase, mannitol fermentation (MSA) (the Merck Group, Germany), and novobiocin susceptibility test. To definitely determine the isolates of S. aureus in cultured samples, the polymerase chain reaction (PCR) assay was performed for the gene encoding deoxyribonuclease (nuc). Following the final identification, the isolates of S. aureus were stored in the culture medium tryptic soy broth (TSB; the Merck Group, Germany) containing 20% glycerol at -70°C. To explore the presence of the methicillin-resistance gene (mecA), the PCR test was performed for all isolates (Table 1).
| Primer | Sequence (5’ - 3’) | Denaturation 94°C | Annealing 45 s | Extension 72°C | Product Size (bp) |
|---|---|---|---|---|---|
| mecA | F: GTAGAAATGACTGAACGTCCGATAA | 45 s | 50 °C | 1 min | 310 |
| R: CCAATTCCACATTGTTTCGGTCTAA | |||||
| intI1 | F: CAGTGGACATAAGCCTGTTC | 45 s | 51 °C | 1 min | 160 |
| R: CCCGACGCATAGACTGTA | |||||
| intI2 | F: TTGCGAGTATCCATAACCTG | 45 s | 51 °C | 1 min | 288 |
| R: TTACCTGCACTGGATTAAGC |
Nucleotide Sequence of Primers Used in the Detection of Integrons in S. aureus (35 Cycles)
3.2. Antibiotic Susceptibility Testing
The antibiotic susceptibility testing was accomplished based on the Kirby-Bauer disc diffusion method at turbidity of 0.5 McFarland standards following the CLSI guidelines (15). We used 11 antibiotic discs (MAST, UK) including gentamicin (10 μg), amikacin (30 μg), tobramycin (10 μg), rifampicin (5 μg), penicillin (5 μg), linezolid (30 μg), vancomycin (30 μg), clindamycin (2 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), and co-trimoxazole (25 μg). S. aureus suspension was spread on Mueller-Hinton agar medium (Himedia Co., India) followed by incubation at 37°C and then compared with a 0.5 McFarland standard using the lawn culture method. Next, the antibiotic disks were placed on the medium. Following incubation in an incubator for 24 hours, the growth inhibition zone diameters were measured and compared with those listed in CLSI tables. To control the quality of the discs, an S. aureus standard strain (ATCC 25923) was used for antibiogram testing and those resistant to at least three antibiotic classes were considered as MDR isolates.
3.3. PCR Assay
A boiling approach was followed to extract the genomes of the isolates. The PCR was similarly performed to detect the genes of class I and II integrons using their specific primers (Takapou Zist Co., Iran) (16) described in Table 1 and with a final volume of 25 μL containing 12.5 μL of Master Mix (SinaClon Co., Iran), 1 μL of each of the primers, 2 μL of bacterial DNA, and sterile distilled water to 25 μL. The PCR thermal cycles (Table 1) for mecA and integron genes included initial denaturation at 94°C for 5 minutes, followed by 35 main cycles and a final extension at 72°C for mecA and integron genes for 2 and 7 minutes, respectively. Shigella flexneri ATCC 12022 and Shigella sonnei ATCC 9290 carrying the integron genes were used as positive controls. Moreover, ATCC 33591 and ATCC 25923 were utilized as positive controls for mecA and nuc genes, respectively. Finally, the PCR products were analyzed by the 2% agarose gel electrophoresis and stained with ethidium bromide.
3.4. Statistical Analysis
The data were analyzed by SPSS software (version 16) using the chi-square test. P values of less than 0.05 were considered statistically significant.
4. Results
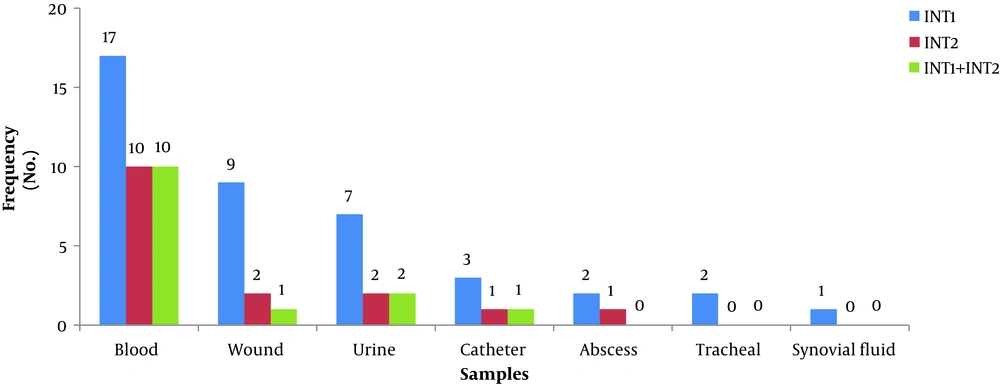
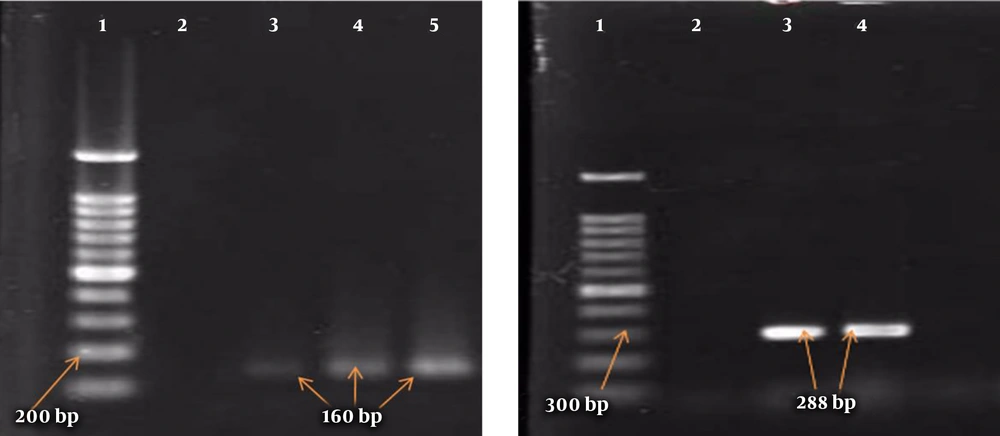
In this study, out of 86 isolates of S. aureus, 54 (62.8%) were found in men and 32 (37.2%) in women, with an overall mean age of 41.80 ± 17.8 years ranging from 7 to 81 years. The highest frequency of the isolates was found in blood samples (n = 28, 32.6%), followed by wound samples (n = 22, 25.6%), urine samples (n = 18, 20.9%), trachea samples (n = 9, 10.5%), catheter samples (n = 4, 4.6%), synovial fluid samples (n = 3, 3.5%), and abscess samples (n = 2, 2.3%). The frequency of MRSA and MSSA isolates was determined to be 50 (58.1%) and 36 (41.9%), respectively (Table 2). In the MRSA isolates, the most antibiotic resistance was observed against penicillin (100%) and gentamicin (80%) and the most sensitivity was reported to vancomycin (100%) and linezolid (94%) (Table 2). Moreover, the rate of MDR isolates was 93.1% (80 isolates). In addition, the frequency of class I and II integrons was 47.7% (41 isolates) and 17.4% (15 isolates), respectively, and 14 isolates had both class I and II integrons. Furthermore, blood and synovial fluid samples had the maximum and minimum frequency of integrons, respectively (Figure 1). Besides, there was a statistically significant relationship between the frequency of class I and II integrons and resistance to antibiotics, especially aminoglycosides (P < 0.05) (Table 3). The PCR results for class I and II integrons are presented in Figure 2.
| Antibiotic | Resistance in MRSA (50 Isolates) | Resistance in MSSA (36 Isolates) |
|---|---|---|
| Gentamicin | 40 (80) | 13 (36.1) |
| Amikacin | 26 (52) | 12 (33.4) |
| Tobramycin | 23 (46) | 5 (13.9) |
| Rifampicin | 24 (48) | 9 (25) |
| Penicillin | 50 (100) | 36 (100) |
| Linezolid | 3 (6) | 0 |
| Ciprofloxacin | 23 (46) | 23 (63.9) |
| Co-trimoxazole | 13 (26) | 6 (16.7) |
| Levofloxacin | 23 (46) | 12 (33.4) |
| Clindamycin | 25 (50) | 13 (36.1) |
| Vancomycin | 0 | 0 |
Results of Antibiotic Resistance of Methicillin-Resistant and Methicillin-Sensitive S. aureus Isolatesa
| Antibiotic | Total Resistance (86 Isolates) | Class I Integron-Positive (n = 41) | Class II Integron-Positive (n = 15) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R | I | S | R | I | S | R | I | S | |
| Gentamicin | 53 (61.6) | 5 (5.8) | 28 (32.6) | 32b | 3 | 6 | 12b | 2 | 1 |
| Amikacin | 38 (44.2) | 1 (1.2) | 47 (54.7) | 22b | 1 | 18 | 9b | 1 | 5 |
| Tobramycin | 28 (32.5) | 3 (3.5) | 55 (64) | 21b | 1 | 19 | 11b | 0 | 4 |
| Rifampicin | 33 (38.4) | 0 | 53 (61.6) | 21b | 0 | 20 | 12b | 0 | 3 |
| Penicillin | 86 (100) | 0 | 0 | 41 | 0 | 0 | 15 | 0 | 0 |
| Linezolid | 3 (3.5) | 0 | 83 (96.5) | 3 | 0 | 38 | 3b | 0 | 12 |
| Ciprofloxacin | 46 (53.5) | 3 (3.5) | 37 (43) | 17 | 3 | 21 | 8 | 1 | 6 |
| Co-trimoxazole | 19 (22.1) | 0 | 67 (77.9) | 11 | 0 | 30 | 3 | 0 | 12 |
| Levofloxacin | 35 (40.7) | 3 (3.5) | 48 (55.8) | 20 | 2 | 19 | 7 | 0 | 8 |
| Clindamycin | 38 (44.2) | 5 (5.8) | 43 (50) | 23b | 3 | 15 | 7 | 1 | 7 |
| Vancomycin | 0 | 0 | 86 (100) | 0 | 0 | 41 | 0 | 0 | 15 |
Association Between Resistance to Antibiotics and the Presence of Integrons in S. aureusa
5. Discussion
The horizontal transfer of resistance genes via integrons is one of the main routes of antibiotic resistance. The integrons are MGEs carrying gene cassettes that can spread the isolates of MDR and subsequently restrict treatment options to control bacterial infections (4, 17). The presence of these elements, especially class I integrons, is one of the reasons for the emergence of MDR isolates (18). The frequency of class I and II integrons in this study was found as 47.7% and 17.4%, respectively. The results of other investigations also indicated the higher frequency of class I than class II integrons (4, 11), which are in line with the findings of the present study. Mostafa et al. similarly reported the high frequency of class I (72.6%) than class II (35.2%) integrons (4). In another study, the frequency of class I and II integrons was reported to be 92.68% and 7.31%, respectively (16). In two investigations conducted abroad, the frequency of class I integrons was reported as 56% and 42.5% (11, 19). There was also a statistically significant correlation between integron-positive isolates and resistance to certain antibiotics, especially aminoglycosides, rifampin, and clindamycin, which were similar to the results of other investigations (4, 16). In a study in China in 2018, all integron-positive S. aureus isolates were resistant to aminoglycoside (20). Besides, Xu et al. observed a relationship between the presence of class I integron and resistance to gentamicin, erythromycin, tetracycline, and cotrimoxazole (21), indicating the presence of different gene cassettes on integrons and thus the involvement of integrons in the occurrence of antibiotic resistance in these isolates. The available results demonstrated the role of these MGEs in the transfer and spreading of various drug resistance patterns (22). Nowadays, the high prevalence of antibiotic resistance among bacterial pathogens is one of the most important public healthcare concerns. In our study, all S. aureus isolates were resistant to penicillin (100%), which was similar to the results of other investigations (23-25). Besides, there was a high resistance rate to gentamicin (80%), amikacin (52%), and clindamycin (50%). The rates of resistance were significantly higher in MRSA isolates than in MSSA isolates, especially to aminoglycosides and clindamycin. Moreover, Safari et al. underlined the high levels of resistance in S. aureus isolates to gentamicin, ciprofloxacin, and clindamycin, which was consistent with the results of the present study (16). The rates of resistance to gentamicin (90.5%), clindamycin (87.5%), rifampin (71.8%), tobramycin, and ciprofloxacin (84.3%) in S. aureus isolates were higher in another investigation (26) than in the present study. The highest susceptibility of isolates was to vancomycin (100%) and linezolid (96.5%). In most investigations, similar to the present study, all S. aureus isolates were reported as sensitive to vancomycin (12, 26, 27). In this study, 3.5% of the isolates were resistant to linezolid. In an investigation by Goudarzi et al., none of the S. aureus isolates was resistant to this antibiotic (12), but Mostafa et al. reported that 17.3% of the isolates were resistant to linezolid (4). In a study by Poorabbas et al., the lowest resistance rates were to cotrimoxazole and gentamicin (42%), ciprofloxacin (34%), clindamycin (24%), and rifampin (10%) while in the present study, the lowest resistance was to vancomycin (0%) and linezolid (2%) (10). In various investigations, the frequency of MDR S. aureus isolates was reported from 75.8% to 100% (6, 23, 28), which was in agreement with the results of this study with a prevalence of 93.1% in our isolates. In general, the antibiotic resistance rate of S. aureus isolates to some antibiotics in this study was lower than that reported in some other investigations, which could be due to regional differences, the low number of MRSA isolates, the type of samples examined, and differences in the consumption patterns of antibiotics, among others (25-27).
5.1. Conclusions
One of the limitations of this study was that we did not assess the frequency of potential gene cassettes on integrons. Given the frequency of the integrons among resistant strains of S. aureus and the risk of rapid transfer of these agents in these isolates, it is necessary to identify isolates with integrons and their relationships with antibiotic resistance patterns, monitor the resistance patterns, and select appropriate antibiotics using the results of phenotypic and genotypic resistance measurements taken by hospital laboratories, which are effective in reducing antibiotic resistance.