1. Background
Tuberculosis (TB) is a re-emerging disease that is still a major global health concern (1, 2). According to the World Health Organization (WHO), a worldwide incidence of 10 million new TB cases and 1.5 million deaths in 2018 have been reported (2).
Tuberculosis is an infectious disease and vaccination is one of the best schemes in preventing infectious diseases. Since the 1920s, Bacilli Calmette-Guerin (BCG) has been the only licensed TB vaccine used for the prevention of TB among children and adults. However, according to numerous clinical trials, this vaccine has some disadvantages, including lack of efficient long-term immunity against TB, various efficacy in adult pulmonary TB (0% - 80%), and side-effects such as severe cutaneous reaction or even disseminated BCG infection, and it is not recommended for HIV-infected individuals. Therefore, more research is required to introduce new-generation TB vaccines (1, 3-5).
M. tuberculosis is a facultative intracellular bacterium which could survive and cause persistent infection within alveolar macrophages using about 1,500 various proteins such as ESAT-6, CFP-10, MTP64, hspX, and Ag85 complex (6).
ESAT-6 (Rv3875) and fibronectin binding protein b (Ag85Bs) (Rv1886c) are known as major virulence factors expressed and secreted in early phases of infection with M. tuberculosis (MTB). These antigens induce strong immune responses both in animals (e.g., IFN-γ or IL-2) and humans (6-8). Several favorable vaccine categories have been designed using ESAT-6 and Ag85Bs proteins, which could induce a high level of immunity against the early stage of MTB infection with valid and impressive results both in animal studies and I, IIa, and IIb clinical trials (5, 6). Recently, epitope-based vaccines have become more popular than classical DNA vaccines. The epitope-based vaccine technology has provided several advantages such as the insertion of multi-immunodominant epitopes from several proteins in the form of one macromolecule, reduction of potential allergenicity, and decreasing cloning and expression problems (9, 10).
Surprisingly, clinical experiments have shown that binding these antigens to the Fc domain of immunoglobulin (Ig) can lead to their selective uptake. The Fc delivery system plays a pivotal role in improving antigen delivery to macrophages and DC cells (11). Consequently, the use of this system leads to efficient uptake of antigens and processing by antigen presenting cells (APCs) and creates great immune response by Th1 and cytotoxic T-cells (CTLs) (11, 12). In addition, subunit vaccines connected to the Fc fragment of IgG increase serum half-life, solubility, stability, and purification of these subunit antigens (11-13).
In recent decades, vaccine design has focused on producing multi-epitope vaccines as a new approach, and research in bioinformatics can help design effective subunit vaccine candidates against TB. Computational tools, which are cost-effective and time-saving, play an important role in detecting appropriate epitopes (14).
2. Objectives
In the current study, based on the early secretory protein of MTB infection, we designed a novel multi-epitope subunit vaccine by focusing on the Fc fragment of IgG using computational tools.
3. Methods
3.1. Selection of Appropriate Targets
There are numerous antigen candidates to be selected in early replication (e.g., EsxA, EsxB, Ag85 complex, and Mpt64) and dormancy (e.g., DosR families and HspX) phases. In this study, we attempted to design a multi-epitope vaccine based on the early phase of MTB infection. These antigens should be immunodominant in pathogenesis and secreted in the early stages of MTB replication.
3.2. Primary Sequence Alignment and Homology
ESAT-6 and Ag85B gene sequences were retrieved from NCBI (www.ncbi.nlm.nih.gov) for in silico analysis and were then compared with other related sequences using BLAST software to select their conserved regions. In addition, the mouse FC fragment of IgG2a (Fcγ2a) domain (coding regions of hinge-CH2-CH3) was added to the final fusion construct to increase the selective uptake process (11, 13).
3.3. Evaluating Physicochemical Features
Online servers, including Uniprot, PDB, and ProtParam were employed to evaluate Ag85B and ESAT-6 antigens. By the use of online servers, some properties (e.g., molecular weight, isoelectric pH, negatively and positively charged residues, half-life and instability index) of these antigens were evaluated (11).
3.4. Primary Constructed Synthetic Sequence
Protein sequences of the Ag85B, ESAT-6, and Fcγ2a domains were joined to one another by GGSSGG and AAY linkers as a synthetic fusion construct, named Ag85b:ESAT-6:Fcγ2a (11).
3.5. Immune-Informatics Analysis
3.5.1. Prediction of MHC class I and II Binding Epitopes
We used IEDB, nHLAPred, and MHC2Pred online tools in order to predict MHC class I and II epitope-restricted binding sequences (15).
3.5.2. Prediction of B Cell Binding Epitopes
Prediction of B cell binding-epitopes was confirmed using the BepiPred online server from the IEDB online tool according to the Kolaskar and Tongaonkar antigenicity scale (15).
3.5.3. Evaluating Epitopes for HLAs Binding
Binding sites of human HLAs, that is, MHC class I and II-restricted epitopes were evaluated using the IDEB online server.
3.6. Fusion Construct Design and Analysis
3.6.1. Primary Synthetic Multi-Epitopes Construct Design
A new mouse construct was designed based on combining T cell and B cell high immune-modulatory of ESAT-6 and Ag85B selected epitopes. These epitopes bind with each other with appropriate linkers, and subsequently, the fusion construct binds to mFcγ2a (mouse IgG2a) for the enhancement of selective uptake by APCs.
3.6.2. Codon Optimization
The multi-epitopes construct was codon optimized using the JCat tool (http://www.jcat.de) online server.
3.6.3. Construct Sequence Analysis
ProtParam (expasy.org/tools/protparam.html) was employed for the evaluation of amino acid composition and physicochemical properties.
3.6.4. Prediction of Dimer Conservation in Protein Modeling Process
The synthetic construct was developed in dimeric form, and protein modeling was performed using the MODELER tool and protein structure prediction servers such as the (PS)2-v2 and I-TASSER online servers. Then, all the models were assessed according to their scores.
3.6.5. Prediction of mRNA Structure
The secondary structure of mRNA was predicted using the Secondary Structure Web Server (16).
3.6.6. Post-Translational Modification (PTMs) of Multi-Epitopes Construct
Post-translational modifications such as glycosylation, phosphorylation, methylation, and acylation are specifically for eukaryotic cells, while such modifications do not exist in prokaryotic cells. Modification of the new construct may affect the structure of constructs. Therefore, modified structures were predicted by several online servers, which are listed in Table 1 (15, 16).
3.6.7. Prediction of Antigenicity Properties of Designed Construct
VaxiJen was employed to evaluate the antigenicity parameters of our multi-epitopes construct.
3.6.8. Prediction of Allergenicity of Designed Construct
AlgPred and PREAL online servers were applied to predict the allergenicity of our construct.
3.6.9. Docking Stimulation
Docking was carried out by the PatchDock and Xex 8.0 software programs to evaluate potential binding groove of MHC I (PDB ID: 3VCL, 4XXC, and 4O2C) and MHC II molecules (PDB ID: 1BX2, 1AQD, 5JLZ) (17).
4. Results
4.1. Primary Sequence Alignment and Homology
We selected ESAT-6 (NC_000962.3) and Ag85B (AY207396.1) gene sequences and compared them with other related gene sequences of the MTB complex. Using BLAST software, 99% similarity between gene sequences was shown for each of the MTB complex genes. Finally, conserved regions were selected for future evaluation. In addition, the mRNA sequence of IgG2a (accession number: V00798.1) was downloaded from NCBI in order to select coding regions of hinge region and Fc domain of mouse IgG2a.
4.2. Evaluating Physicochemical Features
ESAT-6 (PBD: 1WA8) and Ag85B (UniProt: Q847N4) protein sequences were downloaded from PBD and UniProt and evaluated using ProtParam as an initial screening for determining the first suitable confirmation for the vaccine candidate. These proteins were considered appropriate to be selected as a TB vaccine candidate.
4.3. Primary Constructed Synthetic Sequence
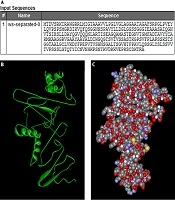
The primary construct was synthesized using the combination of Ag85B-GGSSGG-ESAT-6-AAYAAY-Fcγ2a sequences (Figure 1A-C).
4.4. Immune-Informatics Analysis
4.4.1. Prediction of MHC Class I and II Binding Epitopes
Appropriate immune epitopes were predicted according to BALB/c mouse H-2 class I alleles, including H2-Kd, H2-Dd, and H2-Ld. Three epitopes were selected based on high score values (including LSNRAVKPTGS, MTDVSRKIRAWGRRLMIGTAAAVVLPGLVGLAGGAATAGAFSRPGL, and FSRPGLPVEYLQVPSPSMGR with the total scores of 0.83, 0.81 and 0.71, respectively). Moreover, the protein sequence was submitted on IDEB/MHC II for evaluating potential binding with BALB/c mouse H-2 Class II alleles consisting of H2-IAd and H2-IEd. Subsequently, three epitopes were selected based on the highest score results via online tools, including QGNVTSIHSL, EAGQAMASTEGNVTGMFA, or VQFQSGGNNSPA with the cut-off scores of 0.72, 0.68, and 0.62, respectively.
4.4.2. Prediction of B Cell Binding Epitopes
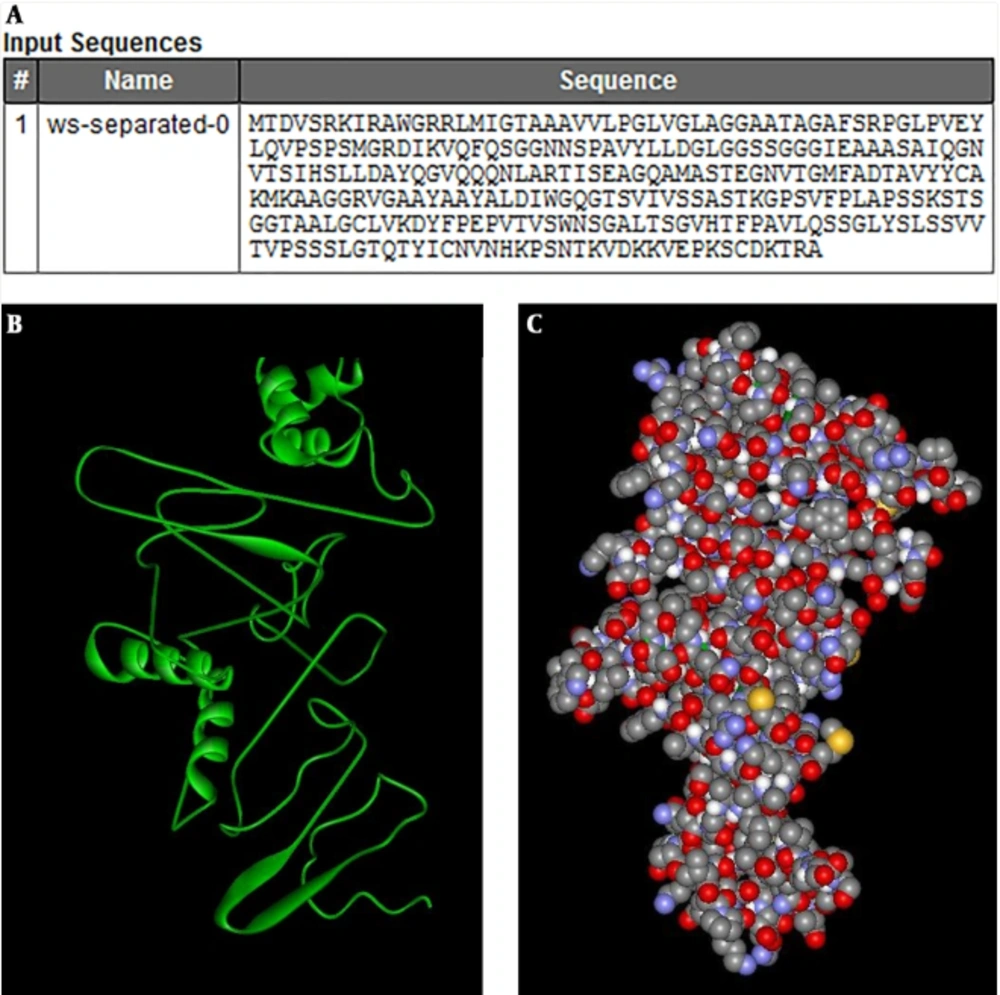
Predicted B cell linear epitopes were AAAVVLPGLVGLA, GLPVEYLQVPS, and AVYLLD domains with an appropriate threshold value higher than 0.5. In addition, Kolaskar and Tongaonkar plot was conducted with a high rate of 1.227 (Figure 2).
4.4.3. Evaluating Epitopes for HLAs Binding
The selected MHC I and II restricted epitopes were analyzed to assess their potential ability in binding to human HLA alleles by the IDEB online tool. All the selected epitopes were confirmed and were able to bind to human HLA alleles with acceptable population coverage higher than 50%, such as AKMKAAGG, GLLGGS, or DKKVEPKS with the total rates of 98.5, 98.0 and 97.5, respectively.
4.5. Fusion Construct Design and Analysis
4.5.1. Primary Synthetic Multi-Epitopes Construct Design
The final construct was designed based on MHC class I, II, and linear B-cell epitopes that were joined using the GGSGG linker and were also connected to the Fcγ2a fragment of IgG (PDB: 1IGT) to increase selective uptake delivery system.
4.5.2. Codon Optimization
The synthetic gene was a codon adapted according to a famous eukaryotic expressing system as Saccharomyces cerevisiae for appropriate post-translational modification process.
4.5.3. Construct Sequences Analysis
Physicochemical parameters of the new construct were evaluated by the ProtParam online tool. The properties of this construct included the number of amino acids: 293, molecular weight: 30 kDa, pH: isoelectric 8.29, negative charge: 20, positive charge: 22, half-life > 30 hours, aliphatic index: 78.57 and gravy: -0.01.
4.5.4. Prediction of Dimer Conservation in the Protein Modeling Process
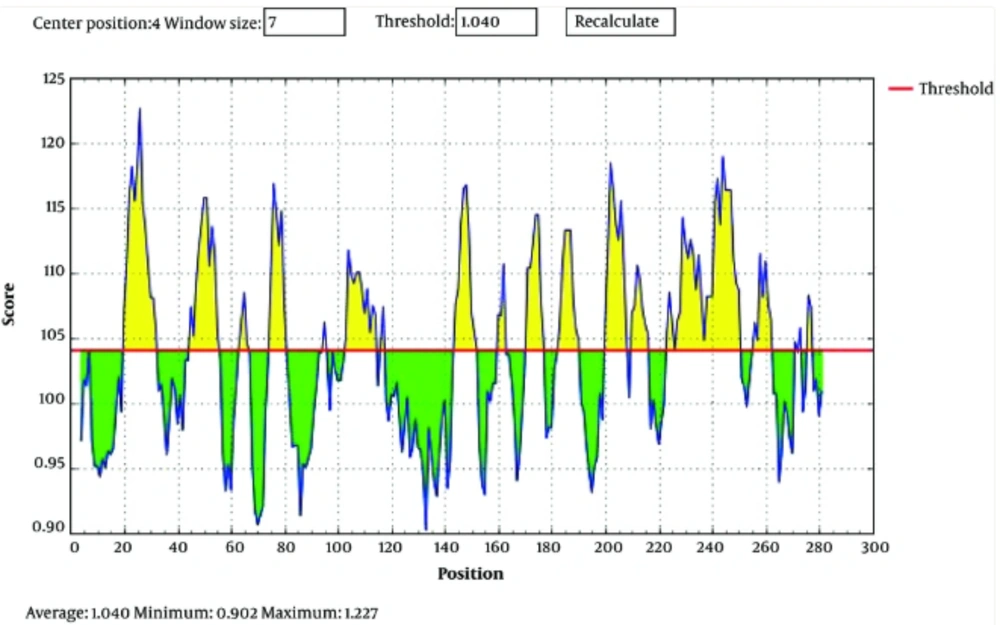
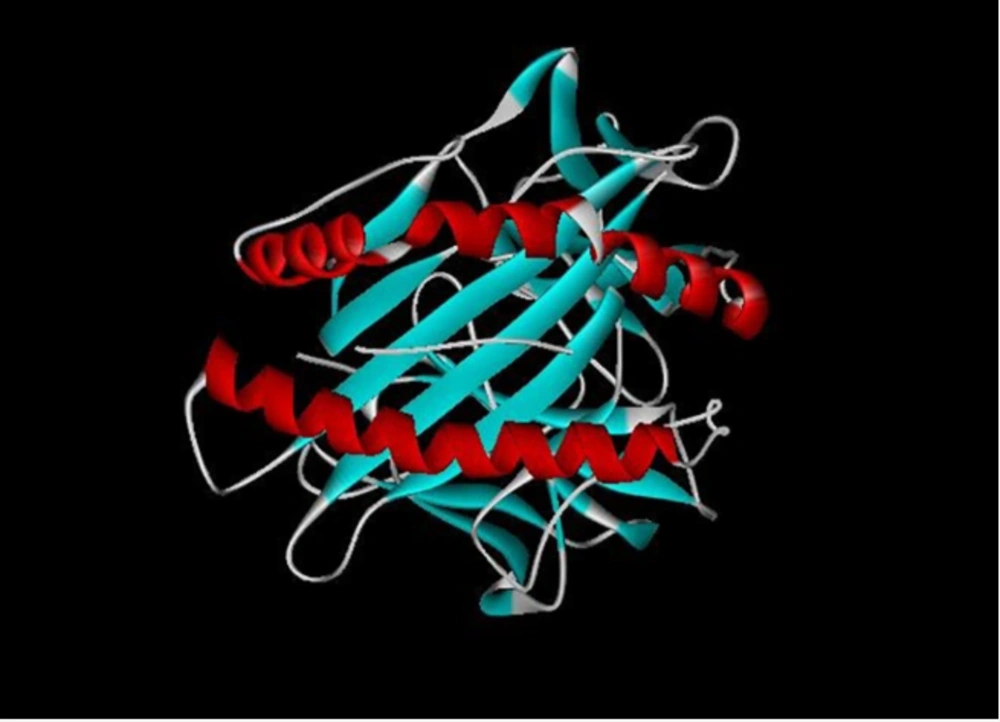
The synthetic gene was converted to dimeric forms, and all the predicted models were evaluated based on a high score rate (GDT_TS score > 60%, ProQ score 1.6, Bit score 120.3 with E value 0.014). Finally, construct stability was confirmed using ERRAT (http://servicesn.mbi.ucla.edu/ERRAT/) and Ramachandran software programs to be 96% and 90%, respectively (Figure 3A-D).
4.5.5. Prediction of mRNA Structure
Twenty mRNA structures were predicted, and all the structures were checked to select the best optimal structure according to ∆G rate. Our mRNA model had ∆G: -225 kcal/mol and did not have leucine zipper or coiled coil, and the hairpin loop and stem loop domain was lower than 20%.
4.5.6. Post-Translational Modification (PTMs) of Multi-Epitope Construct
We employed numerous online servers for PTMs prediction including Glycosciences, NetOGlyc, NetPhos, NetPhosK, Myristoylator, NMT, CELLO, and BaCelLo (Table 1).
| Online-Server URL Address | Function | Modification Types | Residue |
|---|---|---|---|
| http://www.glycosciences.de/ | Prediction of O-linked glycosylation sites in eukaryotes | O-linked glycosylation | T2 |
| www.cbs.dtu.dk/services/NetNGlyc | Prediction of N-linked glycosylation sites in eukaryotes | T13 | |
| http://montana.informatics.indiana.edu/ModPred/index.html; http://montana.informatics.indiana.edu/ModPred/index.html | T15 | ||
| T43 | |||
| T122 | |||
| T202 | |||
| T342 | |||
| T368 | |||
| T516 | |||
| T523 | |||
| T722 | |||
| T762 | |||
| Prediction of di-sulfide bond sites | Disulfide linkage | C14 | |
| C130 | |||
| C198 | |||
| C423 | |||
| C425 | |||
| C429 | |||
| C431 | |||
| www.cbs.dtu.dk/services/NetPhos | Prediction of phosphorylation sites in eukaryote | Phosphorylation | No |
| www.cbs.dtu.dk/services/NetPhosK | Prediction of kinase sites in eukaryote | No | No |
| mendel.imp.ac.at/myristate/SUPLpredictor.html | Prediction of N-terminal myristylation | Lipid | No |
| mendel.imp.ac.at/myristate/SUPLpredictor.html | Prediction of N-terminal N-myristylation | No | |
| www.cbs.dtu.dk/services/SignalP | Prediction of signal peptide (SPase) sites in protein sequence | Signal peptide | E29 |
| gpcr.biocomp.unibo.it/bacello/ | Prediction of extracellular localization of proteins | Protein cellular localization | Extracellular |
| cello.life.nctu.edu.tw/ | Extracellular | ||
| https://wolfpsort.hgc.jp/ | Secreted pathway |
4.5.7. Prediction of Antigenicity Properties of Construct Design
The synthetic gene construct was determined as an antigen according to the VaxiJen online software (cut-off: 0.7).
4.5.8. Allergenicity Evaluation of Construct Design
Both AlgPred and PREAL online tools predicted that the synthetic construct ESAT-6:Ag85B:Fcγ2a was not an allergen (reliability: 90%).
4.5.9. Docking Results
Several epitopes such as SVVTVPSSLGTQTY, FSRPGLPVEYLQVPSPSMGR, VQFQSGGNNSPA and DGLGGSV were evaluated for docking with several MHC class I and II alleles. Among them, SVVTVPSSLGTQTY and FSRPGLPVEYLQVPSPSMGR were bound closely with MHC (4O2C and 1AQD) with the high scores of 11910 and 10196, respectively (Figure 4).
5. Discussion
Nowadays, in addition to therapeutic protocols for the treatment of TB infections, the role of other strategies such as autophagy, antithetical effects of microRNA molecules and the obvious role of T regulatory (T reg) cells in the pathogenesis of TB have been known to researchers (18-21). However, it can be stated that prescription of effective preventive vaccines is considered as an important strategy against TB infection. BCG is the only available TB vaccine approved by the WHO from 1921. However, this vaccine has serious drawbacks and side effects, including its contraindication among HIV patients, its inefficacy in adult pulmonary TB and its inability to control reactivation of the latent form (3, 21). According to the literature, BCG vaccine is not able to induce the production of long-life T cells, which are continuously expressed for pro-inflammatory cytokines. There are numerous cases that have received attenuated BCG vaccine in childhood, but they have been affected by TB after puberty (5). Recently, subunit vaccines have been used as BCG prime boost and are also considered proper candidates for the new generation of TB vaccines. These can provoke CD4+ T cells’ memory, which, in turn, produce a protective response against TB that could remain more than one year (22).
Nowadays, subunit vaccines are applied for several purposes, including prophylaxis (protection), post-exposure (prevention of TB reactivation using dormant antigens) and therapeutic targets (even treatment of MDR-TB) (3, 4). Subunit vaccines such as H1/IC31 can also be recommended for HIV-infected individuals or immunocompromised patients (23). The problems associated with subunit vaccines are high production costs, inefficient bioavailability, low antigen stability during formulation or hydrolysis, and inactivation process (6). Therefore, computational analysis is the first choice for rational design and is considered as the best economical option to predict and process synthetic subunit vaccine properties (16).
Multi-epitope subunit vaccines are new strategies which are developed to produce highly efficient vaccines. The multi-epitope-pulsed dendritic cell vaccine and multi-epitope TARP peptides are famous examples of epitopes which have entered clinical trials (24). Online bioinformatics databases have provided the possibility to predict the antigenic region of proteins. Each of the efficient vaccines should be screened and evaluated by bioinformatics tools before production (14). On the other hand, PCR amplification of high GC-rich genes such as fbpB is challenging and not easily setup. Consequently, construction of fusion epitopes is appropriate and very efficient for triggering cellular immunity response (25). In addition, Ag85B and ESAT-6 are considered as important early expressed antigens of MTB, and subunit vaccine ESAT-6:Ag85B has entered phase IIa of the clinical trial (7).
In the present study, we applied highly immunogenic epitopes of ESAT-6 and Ag85B connected to Fcγ2a and assessed those using certifiable online servers. According to the literature, MTB as a facultative intracellular pathogen is responsible for the activities of the cellular immune system such as Th1 and CTLs control and elimination of TB bacilli from the human body (26). Thus, our construct was designed based on MHC class I and II, T cell, HLA alleles and B cell restricted high score epitopes. Subsequently, fusion-epitopes were combined with the constant region of the Fcγ2a fragment. The Fcγ2a fragment is necessary for enhancing selective uptake and stability and promoting the aliphatic index of the synthetic construct. In order to increase antigenic capacity, multi-epitopes subunit vaccine ESAT-6:Ag85b:Fcγ2a was converted to dimer form, and the 3D model of the dimeric peptide was calculated by I-TASSERS with reliable C-score, ERRAT-score, and acceptable Ramachandran plot.
mRNA stability plays a significant role in the amount of protein expression and total protein stability. Computational analysis showed that the predicted model of our final synthetic construct did not have a hairpin loop or leucine zipper, which in turn, could lead to mRNA stability or prevention of mRNA aggregation, and subsequently, hydrolysis. Moreover, ∆G index is a reliable marker for the prediction of mRNA stability structure, and the low amount of ∆G suggests an increase in mRNA stability (15).
Post-translation modifications cause substantial changes in proteins, which could lead to protein maturation in eukaryotic systems after the completion of the translation phase (27). According to bioinformatics databases, our fusion protein was glycosylated and had disulfide bonds and signal peptides and was expressed as an extracellular protein. According to the literature, glycosylation can improve the immunogenic properties of proteins, and disulfide bonds stabilize the tertiary or quaternary protein structures and fix dimeric peptides together (15, 28). Also, the signal peptide sequence provokes the secretion of proteins outside of the cell.
Docking analysis was evaluated as a reliable test to determine the possibility of interaction of antigens with HLA molecules. The computational docking analysis is reported by energy scores, where low energy scores reveal a high possibility of receptor-ligand interaction.
5.1. Conclusions
In summary, after several screening tests, we concluded that our synthetic multi-epitopes ESAT6:Ag85B:Fcγ2a construct can be considered as a potential vaccine. The reliability of our new construct is established based on various reasons including (1) using ESAT6 and Ag85B as two main secretory antigens in the active phase of MTB infection; (2) using Fc fragment as a natural biological targeted delivery system to APCs; and (3) using these three components in the form of a multi-epitope fusion protein for targeted and strong stimulation of cellular immune system. In addition, basic bioinformatic studies can significantly impact the development of efficient vaccines and reduce production costs. Overall, we hope that this synthetic multi-epitope fusion candidate will be applied for in-vitro and in-vivo studies illin the near future.