1. Background
In nature, bacteria are found in two forms of planktonic and biofilm. Basically, biofilm represents a microbial community in which there are interconnections between different groups. Biofilm can be formed from a different microbial species and on different levels from viable to inert. Different conditions, such as nutrient concentrations, microbial population composition, and hydrodynamic conditions (laminar or turbulent streams), can affect the structure of biofilms. Microorganisms in biofilm communities can perform intercellular actions to adapt to changes in environmental parameters. Many biofilms of bacteria are considered to be major problematic for human’s health, environment and the industry. Biofilms are found everywhere in nature, as mucous membranes on rocks or other objects in water. Bacteria are protected from many harmful agents such as UV light, antibiotics and other antimicrobial agents as long as they are inside biofilms (1).
Approximately, 99.9% of bacteria have the capability to produce biofilms on a broad spectrum of levels such as biological and inanimate levels. Biofilm producing has been reported in the large number of bacterial species such as Enterobacter cloacae, Klebsiella pneumonia, Pseudomonas aeruginosa, Escherichia coli, Staphylococcus epidermidis, Staphylococcus aureus, Haemophilus influenza, Actinomyces israelii, Burkholderia cepacia (2-5).
In the health concept, biofilms cause more than 80% of hospital infections. Biofilms play a role in the development of diseases related to several devices such as central venous catheters, central venous catheter needless connectors, contact lenses, endotracheal tubes, intrauterine devices, mechanical heart valves, pacemakers, peritoneal dialysis catheters, prosthetic joint, tympanostomy tubes, urinary catheters, voice prostheses (6, 7).
Several reasons, including the acquisition of genetic elements associated with antibiotic resistance have been responsible for the resistance of biofilms to antibiotic therapy and have caused some problems for their elimination (8, 9). Therefore, preventing biofilm production and their microbial colonies is a key to successful strategy against infections related to medical equipment (10). Recently, the development of anti-microbial agents and coatings impregnated with substances such as antibiotics, silver, nitric oxide, and newly antimicrobial peptides have been expanding rapidly (11).
However, many of these coatings had cytotoxicity effects. Anti-microbial enzymes are emerging as a new generation of anti-microbial agents. Among these enzymes, glucose oxidase is used to produce H2O2 as a known antimicrobial agent in the food industry. H2O2 is a strong adherent oxidizing agent used in low concentrations as an antiseptic. Cellobiose dehydrogenase (CDH) is an extracellular enzyme produced by various wood-degrading fungi. It oxidizes soluble cellodextrins, mannodextrins and lactose efficiently to their corresponding lactones by a ping-pong mechanism using a wide spectrum of electron acceptors, including quinones, phenoxyradicals, Fe3+, Cu2+ and triiodide ion. Founding an alternative treatment of infections related to biofilm-producing bacteria may be beneficial to clinicians (12-15).
2. Objectives
Among the new technologies based on biotechnology, enzymes are developing as a new generation of antimicrobial agents. Therefore, this study aimed to determine the minimum inhibitory concentration of cellulose dehydrogenase enzyme from Aspergillus niger strains on biofilm of clinical isolates of Staphylococcus epidermidis and Pseudomonas aeruginosa.
3. Methods
3.1. Fungal Strains and Culture
Five standard strains of Aspergillus niger were purchased from the Scientific and Industrial Research Organization of Iran. Each strain was cultured onto Cezapek dox agar (Merk.co1054380500) and incubated at 26ºC and 35ºC until the growth was obtained. The plates were considered during 4 weeks. Aspergillus niger was identified based on colony morphology and microscopic appearance (16).
A volume of 2 - 5 mL distilled water containing 0.1% tween 80 sterile was added to tubes containing fresh colonies of Aspergillus niger. Then, this suspension was divided into sterile 15-mL falcons and the number of fungal cells was counted using Neubauer chamber. (Suspension should be containing 106 cells/mL). Then, dilution of 0.001 of the suspensions was inoculated in Sabouraud dextrose agar containing chloramphenicol and the number of colonies was counted (17).
3.1.1. Zymogram Method for Screening of CDH
To select the strain of Aspergillus niger with the highest enzyme activity in the production of cellobiose dehydrogenase, a solid screening medium was prepared. This medium contained 2, 6-Dichloroindophenol, cellobiose, the Cezapek dox agar, chloramphenicol and cellulose. Twenty μl of spore suspension of fungal strains were inoculated in the well, which was prepared in each plate. Plates were stored at 25ºC for 7 - 10 days. Following glutathione sulfate dehydrogenase production by Aspergillus niger, and the reduction of the 2, 6-DCPIP in the culture, a clear halo was produced around colony that is directly related to the amount of enzyme produced. Therefore, the diameter of the transparent halo formed around each isolate was measured after the incubation period. The isolate that produced the greatest diameter of halo were selected as suitable isolates for the next steps. Fungi selected by this method were used to determine with high enzyme production in a specific culture medium (17).
3.1.2. Enzyme Assay for CDH Activity
The activity of CDH was measured according to Shams-Ghahfarokhi et al. study in 2004. Suitable environmental compositions for measuring the activity of the CDH (cellulose-containing mineral liquid culture medium) were as follows: CoCl2, MnSO4.7H2O, FeSO4.7H2O, ZnSO4.7H2O, CaCl2, yeast extract, 7 H2O MgSO4, KH2PO4, (NH4) 2SO4, cellulose. The amount of 106 fungal cells per mL of culture medium was added in sterile conditions and maintained for 14 days at 28ºC. Mycelia from selected Aspergillus niger were isolated from medium. In order to measure the activity of extracellular CDH, 5 μL of the sample was added to 50 μL 2,6-DCPIP (2 mM), and then 0.9 mL of a 2.5 mM cellobiose solution was added. In order to prepare a control sample, instead of adding a cellobiose solution, a phosphate buffer solution was used and optical density of the samples was read at 600 nm (18).
The blank contained all the above materials except enzyme solution that substituted with an equal amount of phosphate buffer. The reaction was started by the addition of enzyme solution and the decrease in 600 nm absorbance was monitored during the first 5 min at 37ºC. The specific activity was presented as unit/mg protein. Protein concentration was measured by the dye-binding method of Bradford using bovine serum albumin as standard. The final concentration of CDH was 364 U/mL (19).
3.2. Bacterial Strains and Culturing
Bacterial strains were collected from Firoozgar Valiasr Hospital. Only, 42 strains were proper and could produce biofilms. Twenty-four strains of Staphylococcus epidermidis and 18 strains of Pseudomonas aeruginosa were validated with conventional phenotypic and biochemical tests (20). The S. epidermidis strains, ATCC 12228 (American Type Culture Collection) and ATCC 35984 (biofilm-producer) were used (21). Pseudomonas aeruginosa PAO1 (biofilm-producing) and P. aeruginosa ATCC 27853 (non-biofilm-producing) were used as controls (22).
3.2.1. Measurement of Biofilm Production
To investigate CDH ability to consume ExPS as a substrate, overnight cultures of studied strains were grown in TSB at 37ºC. After preparing McFarland concentration, 150 μL of this suspension was poured into 96-well microplates and placed at 37ºC during 24 hours. Then, the supernatant was removed and the wells were gently washed 3 times with physiological saline (0.9 % NaCl) and the microplate was reversed at 65ºC to dry. Better biofilm was used for 96% ethanol at 100 μL. 100 μL ethanol 96% was added, then after 15 min, the alcohol was removed and the microplate was dried in air. Subsequently, 150 μL of 2% crystal violet was added to all wells and after 20 minutes was washed and excess materials were removed from the wells by tapping. Then, 150 μL of 33% acetic acid was added to the wells to release the bonded color to the biofilm to the microplate surface. Optical absorption (OD) was read at 550 nm wavelength using ELISA reader (23).
3.2.2. Antibiofilm Activities of CDH by Microtiter Plate (MTP) Assay
Quantitative biofilm detachment in vitro quantification of biofilm formation was monitored by MTP assay using crystal violet dye and measured spectrophotometrically. Crystal violet dye not only stains but also screens a very small amount of adhered molecules that alter biofilm formation. After 12 h treatment, strains exhibited a reduction in biofilm formation when compared to control. The apparent 50% inhibitory concentration (IC50) of cellobiose in liquid medium was determined by incubating different cellobiose concentrations with Pseudomonas aeruginosa and Staphylococcus epidermidis in 96-well plates. Trypticase soy broth (TSB) (100 µL) was pipetted into each well, followed by 100 µL of 50 mM cellobiose. Serial dilutions of cellobiose ranging from 50, 25, 12.5, 6.125, 3.175, 1.56, 0.078 mM were achieved by transferring 100 µL in each consecutive lane. In the end, a bacterial suspension (Pseudomonas aeruginosa and Staphylococcus epidermidis) containing 5 µL of 105 CFU/mL was added, followed by 10 µL of 364 U/mL CDH. The respective controls, e.g. CDH only, negative and sterile control, were also prepared in the other lanes. Plates were incubated at 37ºC for 24 h and the OD 520 was measured every 30 min in a plate reader. The apparent IC50 value was defined as the concentration of cellobiose inhibiting growth by 50% compared with the negative control. All determinations were performed in triplicate (23, 24).
4. Results
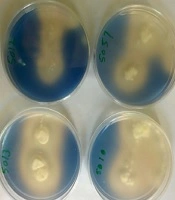
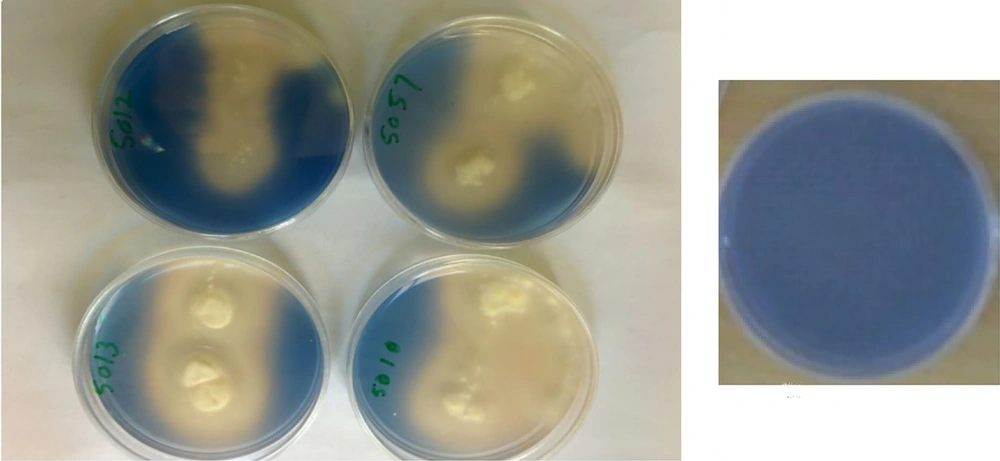
Out of the 42 bacteria gathered from Firoozgar Vali Asr Hospital during June 2017 - 2018, which 24 strains were Staphylococcus epidermidis (57%) and 18 strains were Pseudomonas aeruginosa (43%). There were isolated from blood. In order to select Aspergillus niger CDH producer, all 5 isolates were examined in a 2, 6-dichlorophenolindophenol (DCPIP) screening medium. When DCPIP was oxidized, blue color around fungal colonies was appeared (Figure 1).
Based on zymogram test, isolate No. 5010 was selected as the best isolate of Aspergillus niger for production of extracellular CDH enzymes and used in subsequent stages of the study.
The activity of CDH was measured and CDH activity with 364 U/mg final concentration with a standard deviation were obtained. As it is indicated in Table 1, the results of biofilm formation were classified as biofilm-negative (-), weakly (+), moderately (++), and strongly adherent (+++). Mean and standard deviation based on the strength of biofilm formation in microtiter plate is shown in Table 2 (25).
| Biofilm Class | Status | Results |
|---|---|---|
| If OD ≤ OD c | Non-adherent | (-) |
| If OD c < OD ≤ 2x OD c | Weakly adherent | (+) |
| If 2x OD c < OD ≤ 4x OD c | Moderately adherent | (++) |
| If 4x OD c < OD | Strongly adherent | (+++) |
Classification of Bacteria Based on the Strength of Biofilm Formation in Microtiter Plate
| Strain Number | Mean ± SD | Biofilm Mode |
|---|---|---|
| S.1 | 0.811 ± 0.028 | Moderately adherent |
| S.2 | 0.556 ± 0.028 | Moderately adherent |
| S.3 | 0.621 ± 0.028 | Moderately adherent |
| S.4 | 0.567 ± 0.028 | Moderately adherent |
| S.5 | 0.908 ± 0.028 | Moderately adherent |
| S.6 | 1.08 ± 0.028 | Strongly adherent |
| S.7 | 0.631 ± 0.028 | Moderately adherent |
| S.8 | 0.528 ± 0.028 | Moderately adherent |
| S.9 | 0.564 ± 0.028 | Moderately adherent |
| S.15 | 0.738 ± 0.028 | Moderately adherent |
| S.16 | 0.721 ± 0.028 | Moderately adherent |
| S.17 | 0.612 ± 0.028 | Moderately adherent |
| S.18 | 0.740 ± 0.028 | Moderately adherent |
| S.19 | 0.811 ± 0.028 | Moderately adherent |
| S.20 | 1.253 ± 0.028 | Strongly adherent |
| S.21 | 1.214 ± 0.028 | Strongly adherent |
| S.22 | 0.621 ± 0.028 | Moderately adherent |
| S.23 | 0.438 ± 0.028 | Weakly adherent |
| S.24 | 0.52 ± 0.028 | Weakly adherent |
| S.25 | 0.419 ± 0.028 | Weakly adherent |
| S.26 | 0.480 ± 0.028 | Weakly adherent |
| S.28 | 0.434 ± 0.028 | Weakly adherent |
| S.29 | 0.335 ± 0.028 | Weakly adherent |
| S.30 | 0.429 ± 0.028 | Weakly adherent |
| P.10 | 0.485 ± 0.041 | Weakly adherent |
| P.11 | 0.975 ± 0.041 | Strongly adherent |
| P.12 | 0.608 ± 0.041 | Moderately adherent |
| P.13 | 0.523 ± 0.041 | Moderately adherent |
| P.14 | 0.447 ± 0.041 | Weakly adherent |
| P.27 | 0.531 ± 0.041 | Weakly adherent |
| P.31 | 0.618 ± 0.041 | Moderately adherent |
| P.32 | 1.004 ± 0.041 | Strongly adherent |
| P.33 | 0.468 ± 0.041 | Weakly adherent |
| P.34 | 0.63 ± 0.041 | Moderately adherent |
| P.35 | 0.494 ± 0.041 | Weakly adherent |
| P.36 | 0.52 ± 0.041 | Weakly adherent |
| P.37 | 1.126 ± 0.041 | Strongly adherent |
| P.38 | 0.566 ± 0.041 | Weakly adherent |
| P.39 | 0.490 ± 0.041 | Weakly adherent |
| P.40 | 0.980 ± 0.041 | Moderately adherent |
| P.41 | 0.497 ± 0.041 | Weakly adherent |
| P.42 | 1.140 ± 0.041 | Strongly adherent |
Mean and Standard Deviation Based on the Strength of Biofilm Formation in Microtiter Plate Assay (MTP)
Mean and standard deviation about the strength of biofilm formation in microtiter plate assay, in Pseudomonas aeruginosa strains was 0.672 ± 0.246 and in Staphylococcus epidermidis strains 0.668 ± 0.245, which was not statistically significant (P = 0.955). Classification of bacteria based on the strength of the biofilm formation status in microtiter plate assay was 58.3 % moderately adherent in Staphylococcus epidermidis and 27.8% in Pseudomonas aeruginosa strains, which was not statistically significant based on t-test (P = 0.955).
Of the 42 bacterial samples, 24 strains were Staphylococcus epidermidis, 18 strains were Pseudomonas aeruginosa. Out of 24 bacterial strains of Staphylococcus epidermidis, 7 strains were weakly adherent (29%), 14 strains moderately adherent (58%), and 3 strains strongly adherent (13%). Out of 18 pseudomonas aeruginosa strains, 9 strains were weakly adherent (50%), 4 strains moderately adherent (23%) and 5 strains strongly adherent (27%).
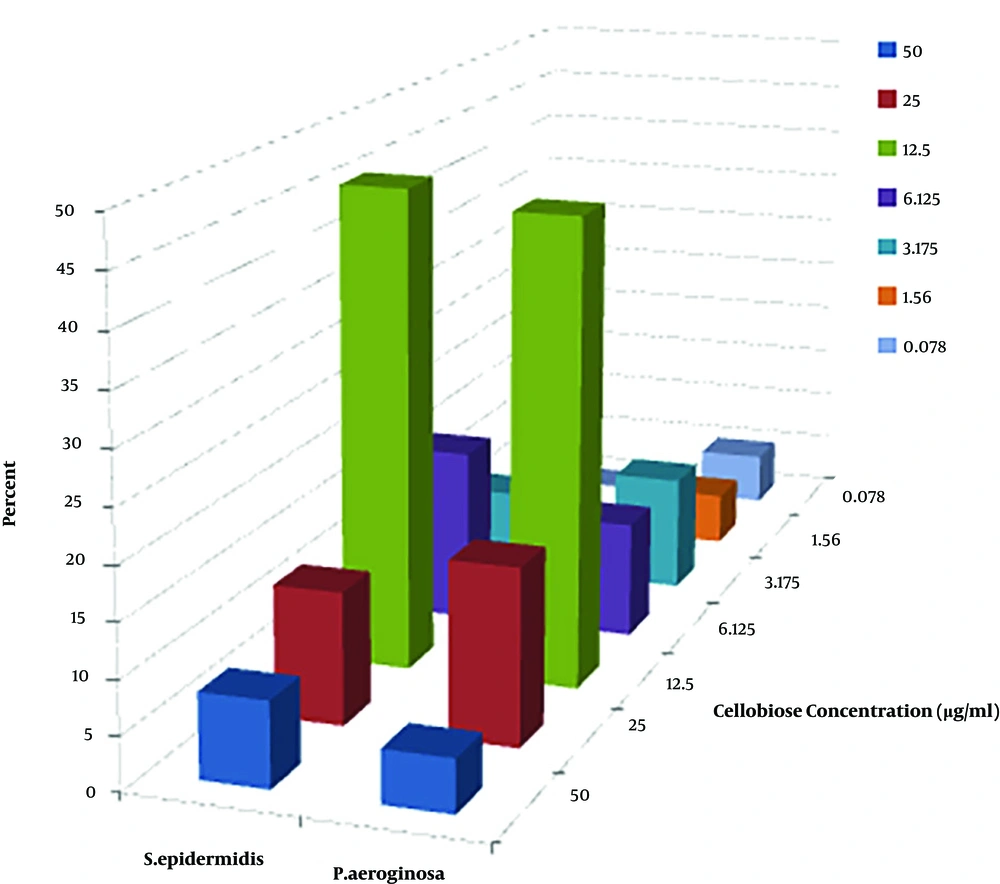
Growth inhibition of Staphylococcus epidermidis and Pseudomonas aeruginosa in liquid cultures as a function of cellobiose concentration in the presence of CDH was considered. Out of 42 strains, CDH had inhibitory concentration only on 34 strains (Figure 2).
Out of 24 pseudomonas aeruginosa strains, 4 strains (22.2%) and out of 18 Staphylococcus epidermidis 4 strains (16.7%) did not have any minimum inhibitory concentration. Fisher exact test revealed that there were no significant differences between pseudomonas aeruginosa strains and Staphylococcus epidermidis (P = 0.71).
In fact, IC50 is a concentration in which 50% of growth is prevented by CDH compared with negative control. According to the results of IC50 shown in Figure 2, the most effective dilution of cellobiose on growth inhibition of Staphylococcus epidermidis and Pseudomonas aeruginosa in liquid cultures as a function of cellobiose concentration in the presence of cellobiose dehydrogenase was in 12.5 µg/mL.
5. Discussion
The bacterial cell wall has a shape-giving function and protects the cells from osmotic disruption. In addition, several antimicrobial agents fail to penetrate the biofilms mainly due to the presence of ExPS, which act as a barrier and protect the bacterial cells. Owing to the heterogeneous nature of ExPS, a mixture of enzymes might be necessary for efficient degradation of bacterial biofilms. Previous reports have shown that enzymes’ mixture can degrade the ExPS of the bacterial biofilms. Enzymes degrade the biofilms directly by destroying the physical integrity of the extracellular polymeric substances through weakening the proteins, carbohydrate and lipid components of the extracellular polymeric substances (8, 26-30).
Microbial biofilms are now a serious medical problem. Because biofilms have become a major concern today, and bacteria indicate strong resistance to antibiotics and other disinfectants through these structures. Several antimicrobial enzymes targeting different cellular components and biofilms are intensively being investigated with some products already commercialized in the health, food and biomedical industry. Although generally, enzymes are effective as antimicrobial agents, successful removal of complex biofilms requires the use of a complex enzyme formulation containing DNase to degrade extracellular DNA, CDH extracellular polysaccharides, proteases to hydrolyze proteins as well as anti-quorum sensing enzymes to prevent biofilm formation. A novel in situ antibiofilm and antimicrobial system based on the ability of CDH to produce H2O2 in the presence of cellobiose was successfully developed (14).
The first and only report on the production of this enzyme in the Aspergillus niger species of was reported by Duarte et al. in 1999. The researcher showed that by using the zymogram method, different isolates of different fungal species, including different species of Aspergillus niger, could be studied in terms of the production of CDH enzymes. They showed that the CDH enzyme in the Aspergillus niger fungus is an extracellular enzyme and is secreted into the culture medium (31).
We determined MIC of CDH enzyme extracted from Aspergillus niger on the biofilm of Staphylococcus epidermidis and Pseudomonas aeruginosa isolated from clinical specimens.
Croes et al. in 2009, studied Staphylococcus aureus biofilm formation at the physiologic glucose concentration based on the S. aureus lineage. They found the adherence to polystyrene surfaces under physiologic glucose concentration (0.1%) was dependent on the clonal lineage (32). In fact, biofilm inhibition more closely correlated with increasing substrate concentration. When the concentration of the substrate increased, the biofilm inhibition decreased. In our study, we reported the highest concentration of cellobiose was in 12.5 µg/mL. We also found that increasing biofilm inhibition was associated with increasing substrate concentrations. When the concentration of the substrate is greater than 12.5 µg/mL, biofilm was reduced. We also found that inhibition of CDH was increased by the presence of cellobiose as a substrate.
CDH/cellobiose dilution was able to oxidize enzymatically ExPS of Staphylococcus epidermidis and Pseudomonas aeruginosa leading to the production of H2O2. These complexes of CDH/cellobiose were known to exhibit antimicrobial activities by destroying the biofilm matrix through linked glycosides cleavage of ExPS. Hydrolysis of polysaccharides by this enzyme increases the number of terminal reducing sugars as substrates for CDH as well as destabilizes the biofilm. In addition, biofilm hydrolysis leads to its destabilization and failure to protect microorganisms from antimicrobial agents (14, 33, 34).
An increase of biofilm inhibition with the increase of the substrate for CDH was expected. The inhibition of biofilm formation was decreased at a substrate concentration of 6.25 µg/mL cellobiose. The reason could be explained by the metabolization of cellobiose by the cells which were embedded in the bacterial biofilms was faster than the enzymatical reaction (34).
5.1. Conclusions
Based on the results of this study, it can be concluded that the CDH enzyme can be a candidate as an anti-microbial agent. Interestingly, CDH was also able to produce H2O2 during oxidation of ExPS formed by microorganisms in cellobiose absence. To develop the antimicrobial system for application further studies should be carried out to exclude the effect of the CDH substrates to support the growth of microorganisms.