1. Background
Candida albicans can cause a variety of infections, ranging from superficial mucocutaneous candidiasis to systemic infections that involve major organs including the lungs, kidneys, liver, spleen, heart, and other organs (1). Recently, with the increasing number of immune-compromised populations, the incidence of candidiasis has also remarkably increased (2).
It has recently documented that molecular factors play significant roles in the pathogenesis of Candida albicans in various anatomical sites of the host (3). A set of virulence factors are involved in C. albicans infection including hyphae formation, phenotype switching, biological substrates’ adhesion, and extracellular production of hydrolytic enzymes (4).
Previous research found that alternation in the mRNA expression level of specific Secreted Aspartyl Proteinase (SAP) genes was related to the active form of the disease and the anatomical site of infection (5). The first step of Candida infection is the formation of hyphae and adhesion to host cells. It seems that the filamentous form is more invasive than the yeast form to the progression of Candida infection (6). Candidal cell adherence to host tissues is a complex multifactorial phenomenon. A 34-kDa adhesin molecule named HWP1, which is found in the hyphal wall protein, is a gene required for virulence in mucocutaneous candidiasis because it encodes an outer surface mannoprotein (7). Hypha-specific expression of HWP1 results in the increased fungal growth in the host in candidiasis in the murine stomach (8). The contribution of HWP1 to mucosal symptomatic and asymptomatic infections has been reported (1).
The most important hydrolytic enzymes produced by C. albicans include Phospholipases (PLBs) and Secreted Aspartyl Proteinases (SAPs) (9). Phospholipase A, B, C, and D have been recognized in C. albicans. Phospholipases have the properties of both lysophospholipase and hydrolase (7). The PLB1 and PLB2 gene products have been detected extracellularly. An 84-kDa PLB1 glycoprotein isolated from the hyphal tip in the course of tissue invasion has shown to be required for virulence in a murine model of candidiasis (5, 7).
2. Objectives
The present study was designed to answer the hypothesis of whether PLB1 and HWP1 mRNA expression patterns are related to the progression of infection in different anatomical sites of the body.
3. Methods
3.1. Sample Collection and Identification of C. albicans Isolates
An experimental study was performed on 120 clinical isolates of C. albicans obtained from various sites of immune-compromised and non-immune-compromised patients who were admitted to teaching hospitals of Shahid Beheshti University of Medical Sciences, Tehran, Iran from 2016 to 2019 (Table 1).
| Source | Number of Samples |
|---|---|
| BAL | 30 |
| Oral | 30 |
| Vaginal | 30 |
| Cutaneous (nail, axillary, groin) | 30 |
Distribution of Clinical Isolates of Candida albicans
The specimens were submitted to the Medical Mycology Laboratory of the Medical School. Initially, all samples were cultured on Sabouraud-dextrose agar (Merck, Germany) at 32°C for 48 hours. Then, a single colony was subcultured on CHROMagar Candida medium (CHROMagar, France), followed by incubation at 35°C for 48 hours to obtain a pure colony of yeasts. A single colony of all isolates was subjected to HWP1 gene amplification to discriminate against the C. albicans complex.
3.2. Amplification of HWP1 Gene
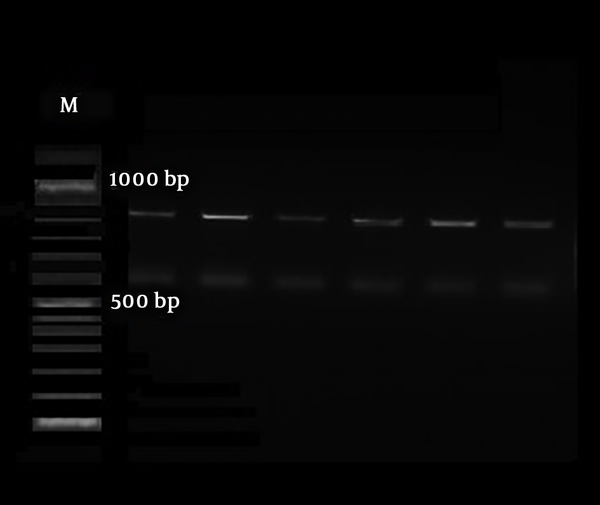
The HWP1 gene was amplified to differentiate the C. albicans complex comprising C. albicans, C. Africana, and C. dubliniensis. The PCR with paired primers HWP1-F (5′-GCTACCACTTCAGAATCATCATC-3′) and HWP1-R (5′-GCACCTTCAGTCGTAGAGACG- 3′) was done in the following condition: 95°C for 15 minutes; 35 cycles of 30 seconds at 95°C, annealing for 30 seconds at 60°C, one minute at 72°C, and a final extension of 5 minutes at 72°C. The data were interpreted according to bands sizes of ~900 bp, ~700 bp, and ~ 560 bp for C. albicans, C. Africana, and C. dubliniensis, respectively. Five microliters of PCR products were electrophoresed on a 1.5% agarose gel in TBE buffer (Merck, Germany); then, they were observed and photographed under ultraviolet irradiation.
3.3. RNA Extraction
All C. albicans strains were cultured on Sabouraud Dextrose Agar (Merck, Germany) and incubated for two days at 35°C. Total RNA was extracted from an exponential phase using the commercial kit (Fermentas, EU) (10, 11). The total RNA was treated using the DNase1 (Fermentas, USA) to improve the reliability of the quantitative method.
3.4. cDNA Synthesis
Single-strand complementary DNA (ScDNA) was synthesized using 2 μg of total RNA in a 10-μL reaction mixture using the Prime Script RT reaction kit (Takara, Japan) according to the manufacturer’s instruction (12). The accuracy of ScDNA was checked with actin (ACT1) gene primers as the housekeeping gene. The PCR condition was as follows: initial denaturation for 5 minutes at 94°C, 30 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 60°C, extention for 45 seconds at 72°C, with a final extension for one minute at 72°C.
3.5. Quantitative Real-Time PCR Assay
The set of primers used the Oligo Explorer (version 15) software. Each primer pair was aligned with sequences from the NCBI GenBank database using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) to ensure the specificity of the sequences. The primers used in this study were synthesized by Bioneer Company (Korea) as listed in Table 2. Quantitative real-time PCR (QRT) was performed with Applied Biosystems 7300 Real-Time PCR System (Singapore). SYBR Premix Ex Taq II (Amplicon, Korea) was used as a reagent specifically designed for intercalator-based real-time PCR. The PCR mixture contained 2 μL of the first-strand cDNA, 10 μL of SYBR green, 0.8 μL of each primer, 0.4 μL of Rox reference dye (50x), and 6 μL of dH2O to make a final volume of 20 μL. The PCR was performed with a preliminary hold at 95°C for 30 seconds as the initial denaturation step, followed by 40 cycles of PCR consisting of 95°C for 5 seconds and 60°C for 31 seconds. Final holding was performed at 72°C for 60 seconds.
The ACT1 gene was used as a housekeeping gene to normalize the data. For the accuracy of the examination, we measured the PLB1 and HWP1 mRNA expression levels of control strains (including reference strain and clinical strain isolated from patients with no invasive candidiasis). The average expression level of them was used as the baseline. All experiments were done in duplicate and each experiment was repeated twice on two different days to assess reproducibility (13).
| Gene | Primer | Sequence |
|---|---|---|
| HWP1 | Forward | 5’ GACCGTCTACCTGTGGGACAGT 3’ |
| Reverse | 5’ GCTCAACTTATTGCTATCGCTTATTACA 3’ | |
| PLB1 | Forward | 5’ GGTGGAGAAGATGGCCAAAA 3’ |
| Reverse | 5’ AGCACTTACGTTACGATGCAACA 3’ | |
| ACT1 | Forward | 5’TGGAGCTTCGGTCAACAAAACTGG 3’ |
| Reverse | 5’ ACGGTATTGTTTCCAACTGGGACG 3’ |
Primers Used in Quantitative Real-time PCR Analysis
3.6. Statistical Analysis
The frequency of the PLB1 and HWP1 genes among C. albicans strains isolated from four clinical sites was analyzed using the Fisher exact test with a significance threshold of P < 0.05. Finally, data obtained from real-time PCR were interpreted using the comparative Ct method (∆∆Ct) using REST© software (2009, V. 2.0.13).
4. Results
4.1. Interpretation of Demographic Information
We collected 120 samples from four clinical sites. Demographic information (age and sex) is shown in Table 3. The immune-compromised patients (n = 80) profile included solid-organ cancers (n = 25), bone marrow transplant recipients (n = 32), and intensive care unit (ICU) patients (n = 23). No patients were treated with antifungal agents at the time of sampling.
| Specimens | Age | Sex | Expression of PLB1 | Expression of HWP1 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Up | Down | Up and Down | Up | Down | Up and Down | ||
| Oral (n = 30) | 46 ± 8.72 | 16 (53.3) | 14 (46.6) | 14 (46.7) | 5 (16.6) | 19 (63.3) | 21 (70) | 9 (30) | 30 (100) |
| Vaginal (n = 30) | 41 ± 10.29 | 30 (100) | 16 (53.3) | 9 (30) | 25 (83.3) | 25 (83.3) | 4 (13.3) | 29 (96.6) | |
| BAL (n = 30) | 47 ± 8.38 | 17 (56.6) | 13 (43.3) | 12 (40) | 8 (26.6) | 20 (66.6) | 13 (43.3) | 10 (33.3) | 23 (76.6) |
| Cutaneous (n = 30) | 42 ± 11.68 | 8 (26.6) | 22 (73.3) | 1 (3.3) | 15 (50) | 16 (53.3) | 9 (30) | 10 (33.3) | 19 (63.3) |
| Total | - | - | 43 (35.8) | 37 (30.8) | 80 (66.6) | 68 (56.6) | 33 (27.5) | 101 (84.1) | |
| P value (between groups) | - | - | - | - | 0.003 | - | - | 0.001 | |
Frequency of Candida albicans PLB1 and HWP1 Genes and Their Expression (Up and Downregulation) in Oral, Vaginal, BAL, and Cutaneous Samplesa
4.2. Result of Amplification of HWP1 Gene
The initially identified C. albicans complex on chromogenic media was discriminated using the amplification of the HWP1 gene. Based on bands revealed on gel electrophoresis, all strains were known as C. albicans (Figure 1) (14).
4.3. Result of QRT PCR
The PLB1 and HWP1 genes were detected in C. albicans strains. The HWP1 gene was detected at a higher frequency than the PLB1 gene in C. albicans strains. There were statistically significant differences in PLB1 and HWP1 gene expressions between C. albicans strains isolated from four clinical sites (P = 0.003 and 0.001, respectively) (Figures 2 and 3).
The HWP1 mRNA expression levels of clinical samples were upregulated by 70%, 83.3%, 43.3%, and 33.3% in oral, vaginal, BAL, and cutaneous samples, respectively (Table 3). The PLB1 mRNA expression level of all samples was upregulated by 46.7%, 53.3%, 40%, and 3.3% (P < 0.001) in oral, vaginal, BAL, and cutaneous samples, respectively, compared to the control group (Table 3).
5. Discussion
In this study, BAL samples from patient specimens had positive blood cultures. Given the significant expansion of Candida spp. infections, as well as differences in pathogenicity, the rapid and accurate identification at the species level seems crucial for clinical management. This study analyzed the mRNA expression levels of the HWP1 and PLB1 genes in C. albicans isolated from 120 oral, vaginal, BAL, and cutaneous samples.
The results provided insights into the patterns of the two genes’ mRNA expression levels in four anatomical sites. In our cases, the expression rates of HWP1 and PLB1 genes were significantly different (P = 0.001 and P = 0.003, respectively) at the four anatomical sites. The results showed 56.6% and 35.8% upregulation in the HWP1 and PLB1 genes, respectively.
The present findings suggest a meaningful correlation between the up-regulation of HWP1 mRNA expression level with the pathogenicity of C. albicans strains and the sites of infection. The data showed that the HWP1 gene could be expressed by C. albicans strains in vitro from the vagina (83.3%). The HWP1 expression was significantly different between vaginal samples from oral and cutaneous samples (P ≤ 0.05) but the difference was not significant between vaginal and BAL samples (P = 1).
The HWP1 mRNA expression is the key player in the activation of vaginal candidiasis and hence, it may have an essential role in the clinical presentation of vaginitis. The HWP1 gene expression was upregulated in mucosal samples compared to cutaneous samples (only 30%). The importance of the HWP1 level in the severity of mucocutaneous candidiasis by C. albicans has been shown previously. The HWP1 gene expression level has been found to affect the pathogenesis of candidiasis in vaginal and oral sites (5). A survey by Nas et al. showed that the expression of HWP1 was detected as 62% in C. albicans strains isolated from women with vulvovaginal candidiasis (10). Pakdel et al. reported that the presence of HWP1 is an important factor for vaginal infection (11).
The results also showed that the PLB1 expression in vaginal samples had a significant difference with the PLB1 expression in cutaneous samples (P ≤ 0.05) while there was no difference with the two other samples (BAL and oral samples). The role of phospholipases in C. albicans infections is also well established through the cloning of a phospholipase gene, PLB1, the deletion of which did not result in alterations in adherence, but generated cells with reduced invasion ability (12).
The phospholipase activity was detected in 35.8% of the C. albicans isolates in the present study. Previous studies have shown phospholipase activity in 30% to 100% of Candida isolates from various groups of patients and various sites (13, 15). The proportion may depend on the site; for example, the phospholipase activity was found in 55%, 50%, and 30% of Candida spp. isolated from blood, wound infection, and urine, respectively (13). The phospholipase gene expression has shown to be affected by growth conditions (15).
In a study by Hoover et al., a little phospholipase activity was detected in C. albicans strains isolated from the oral cavity of carriers (16). The PLB1 expression may be regulated by factors that also regulate the expression of hyphal morphology.
Other possible factors affecting candidiasis should be addressed by evaluating virulence factors other than these genes. If the critical virulence genes were identified in the pathogenesis of candidiasis, it could be an important practical application in developing new diagnostic tests and therapeutic strategies for the treatment of candidiasis (10).
The presence of samples with no expression of HWP1 and PLB1 genes mRNA confirms the recent hypothesis that there is a meaningful relationship between higher expression levels of candidate genes mRNA and the presence of infections in a specific site of the body.
In conclusion, the present study is the only report addressing the expression of the C. albicans HWP1 and PLB1 genes in BAL and cutaneous samples, which demonstrated differences in expression during the course of the disease. Once more, PLB1 and HWP1 were expressed predominantly in mucosal specimens (oral, vaginal, and BAL). This clearly shows that the expression pattern of these candidate genes depends on the organ localization. However, more studies on larger samples are required to characterize the exact molecular mechanism of candidate genes involved in the severity of symptoms, as well as their contribution to the site of infection.