1. Background
The increasing appearance of multidrug-resistant (MDR) microorganisms in clinics is a severe rising threat to human health (1). Most recently, an estimated 2.5 million people acquire antibiotic-resistant infections every year in Europe and the USA leading to approximately 50,000 deaths (2). Methicillin-resistant Staphylococcus aureus (MRSA) is an MDR organism. It was initially detected during the early 1960s in the United Kingdom and is now regarded as a major hospital-acquired pathogen throughout the world (3, 4). Methicillin-resistant S. aureus is one of the most common nosocomial pathogens infecting burn wounds (5). Burn patients are at an increased risk of colonization and subsequent infections by nosocomial pathogens due to the disruption of the skin protective barrier and reduction of immune responses, which can lead to poor clinical outcomes and increased morbidity and mortality rates (6). There exist reports of the outbreaks of MRSA and evidence of increasing MRSA strains in burn units. In addition, reports indicate that burn units and ICUs may act as reservoirs for MRSA. Despite the full compliance with infection control programs, the acquisition and transmission of MRSA are continuous problems in burn units (7).
Staphylococcus aureus is a common commensal bacterium that, as an opportunistic pathogen, is capable of causing a variety of diseases ranging from mild skin infections and food poisoning to fatal infections like endocarditis, pneumonia, osteomyelitis, and toxic shock syndrome. It is also a leading cause of infections associated with catheters and devices (8-11). The pathogenesis of S. aureus strains are related to the expression of different virulence factors, including cell surface components (e.g., collagen-binding protein, clumping factor, fibronectin-binding protein, and elastin binding protein) and secreted factors (e.g., staphylokinase, toxic shock syndrome toxin-1, hemolysin, panton-valentine leukocidin (PVL), exfoliative toxins (eta and etb), staphylococcal enterotoxins (SEs), and lipase) (12).
Unfortunately, MRSA is resistant to all beta-lactam antimicrobial drugs such as penicillin, cefoxitin, and oxacillin, except for newer cephalosporins with anti-MRSA activity. The MRSA isolates are susceptible only to glycopeptides, such as vancomycin (13). In addition to the increasing prevalence of MRSA, the emergence of vancomycin-resistant or vancomycin-intermediate S. aureus (VRSA or VISA) has caused a serious problem (3, 14). Also, community-acquired MRSA infections have increasingly been identified in the past two decades (10). This microorganism has been declared as an international concern by the World Health Organization (1, 15, 16). It is estimated that 70% of mortality in burn units is related to infections. Thus, the management of burn patients is a significant problem because of the outbreaks of infections in burn units and the presence of many MDR strains. Additionally, hospital burn units are the main reservoirs for MRSA with the potential for rapid dissemination in the hospital environment (17). Shahsavan et al. (5) isolated S. aureus strains from burn patients in Tehran and reported that most MRSA isolates were MDR and showed resistance to β-lactams, macrolides, tetracyclines, and aminoglycosides. As a result, therapeutic strategies to encounter clinical infections caused by these bacteria have become limited (2). Therefore, alternative antibacterial agents and programs must be developed (2). Moreover, due to the rapid acquisition of resistance to new antibiotics and rising production costs, there has been little incentive to develop new antibacterial agents (2, 14, 18).
Phage therapy has attracted a great deal of attention as a strategy against MRSA infections and to provide a new solution against the threat of MDR infections (19). This approach uses bacterial viruses (phages) that can specifically attack pathogenic bacteria and kill them (20). Phage therapy has the potential of being highly specific merely against a species or even a strain responsible for infection (8, 21, 22). Moreover, their remarkable specificity prevents them from affecting human cells and the composition of the body microbiota and inducing antimicrobial resistance in different bacterial species (1, 22).
2. Objectives
The aim of the present study was to isolate and characterize a lytic bacteriophage from hospital sewage to be effective against burn wound-infecting MRSA isolates.
3. Methods
3.1. Isolation and Characterization of Bacterial Strains
During a nine-month period starting from September 2017 to May 2018, 30 MRSA isolates were obtained from burn patients hospitalized at Motahari Hospital in Tehran, Iran, and transferred to the Research Laboratory of Microbiology Department, Iran University of Medical Sciences. Strains were identified as S. aureus by Gram staining, coagulase, catalase, oxidase, and DNase production, as well as mannitol fermentation and PCR amplification of the protein A gene (spa). The Kirby-Bauer disk diffusion method was performed to confirm their resistance to penicillin, methicillin, and oxacillin, as recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines. Additionally, PCR assays were carried out for the mecA gene. After the identification of MRSA isolates, they were stored in 20% glycerol at -20°C. Three clinical isolates (MH-1, S1, and M2) plus a reference strain of MRSA (ATCC 43300) were used as hosts for bacteriophage isolation and propagation from wastewater samples.
3.2. Wastewater Sample Collection for Bacteriophage Isolation
Several wastewater samples were taken from untreated sewage ponds of Motahari Hospital and screened as sources of bacteriophages. The samples were centrifuged at 2,500 × g for 20 min to separate bacterial cells and debris. The supernatant was filtered through 0.45 µm filters, transferred to a clean tube, and stored at 4°C (23).
3.3. Isolation and Purification of Bacteriophages
For phage isolation, 10 mL of exponential phase culture of clinical and standard strains of S. aureus was mixed with 10 mL of fresh LB broth (2X) and 10 mL of filtered sewage sample, and incubated overnight at 37°C with shaking at 80 rpm. The culture was then centrifuged for 20 min at 2,500 × g to remove bacteria and the supernatant was filtered through 0.22 µm pore size Millipore filters (for phage amplification, the simultaneous culturing step was repeated three or more times). This lysate (supernatant) was examined for lytic phage in the plaque assay by the Double-layer Agar (DLA) method (23).
3.4. Double Layer Agar
In this method, one milliliter of phage lysate was mixed with 500 µL of a stationary phase culture of host S. aureus. After 10 min incubation at 37°C, 5 mL of LB top agar (4 mM CaCl2, 4 mM MgSO4, 0.7% (w/v) agar, at 45°C) was added, mixed, and overlaid onto fresh LB agar plates (1.5% (w/v) agar) prepared before. After solidifying, the plates were incubated overnight at 37°C until lysis zones appeared (24).
3.5. Purification of Bacteriophage
Single plaques from each plate were picked by a sterile Pasteur pipette and placed in a tube containing one milliliter of LB broth (45°C). One milliliter of stationary phase culture of host bacteria was added to each tube and incubated for 24 h at 37°C and 80 rpm. On the next day, the lysate was screened for the presence of plaques as described above. Purification was carried out by three serial single-plaque isolations (25).
3.6. Determination of Bacteriophage Titer
Ten sterile tubes containing 900 µL Luria Bertani (LB) broth were numbered from 10-1 to 10-10. Next, 100 µL of phage lysate was added to the first tube (10-1), mixed well, and then 100 µL was transferred to the second tube (10-2) in series (10-1 to 10-10). The same pattern was used to make serial dilutions. Then, 100 µL of an exponential phase culture of host MRSA was added to each tube and mixed with 100 µL of each dilution of phage lysate (10-5 to 10-9). Tubes were incubated for 10 min at 37°C for phage adsorption. Five tubes containing 3 mL of LB soft agar (0.4% agar, 4 to 10 mM CaCl2, and MgSO4) at 45°C were numbered from 10-5 to 10-9. The phage and bacterial suspensions were added to soft agar tubes and after mixing were overlaid onto plates containing LB agar (1.5% agar). Plates were incubated overnight at 37°C and the resulting plaques were counted. In this way, PFU/mL was equal to the number of plaques/dilution × volume of diluted phage added to each plate (26).
3.7. Host Range Determination
Thirty MRSA clinical isolates, recovered from burn patients, were used to determine the host range of the isolated bacteriophage. To determine the susceptibility of methicillin-susceptible S. aureus (MSSA) strains to phage-mediated lysis, 30 of its strains from burn patients were also examined. Bacterial strain susceptibilities were detected by the spot test method described by Kutter with some modifications (24). Briefly, bacterial strains were incubated for 4 h in LB broth at 37°C and 180 rpm (OD 600 nm= 0.4 - 0.6). Then, 500 µL of each bacterial culture was added to 6 mL of 0.4% LB soft agar (45°C) and poured onto LB agar plates. The plates were left to dry for 10 min. Subsequently, 10 µL of the phage lysate (109 PFU/mL) was spotted on lawns of different bacterial strains and incubated overnight at 37°C for the formation of a lysis zone. Lytic activity of the isolated phage was also examined on Staphylococcus epidermidis, Enterococcus faecalis, and Enterococcus faecium strains.
3.8. Electron Microscopy
In this method, 10 µL of polyethylene glycol precipitated phage particles were spotted onto a carbon-coated copper acid grid for 3 - 5 min and then blotted with a filter paper and stained with 1% (w/v) uranyl acetate (pH = 7). It was examined by a Zeiss LEO 906 transmission electron microscope (Carl Zeiss LEO EM 906 E, Germany) at an accelerating voltage of 100 kV (26).
3.9. Naming the Isolated Bacteriophage
We named one of our isolated phages as vB-StuM-MH-1 according to the newly proposed naming system vBStuP/M/S MHno, where vB refers to bacterial virus, Stu is an abbreviation for genus/species of the host, P refers to podovirus, M to myovirus, S to siphovirus, and MHno to the name and number of the phage. The last part of the name (MH-1) is the phage’s common name (27).
3.10. One-Step Growth Curve
One-step growth experiments were performed using a method described by Wang et al. (28) with some modifications. Briefly, a mid-exponential-phase culture (30 mL) of S. aureus (OD600nm = 0.4 to 0.5) was harvested by centrifugation and re-suspended in 7.5 mL of fresh LB broth. Phage lysates (109 PFU/mL) were added at an MOI of 0.0005 and incubated at 37°C for 15 min for phage adsorption. The mixture was then centrifuged at 10,000 × g for 10 min to remove free phage particles. The pellet was re-suspended in 10 mL of LB broth and incubated at 37°C. Samples were removed at 10-min intervals for 2 h. The samples were immediately diluted 10-fold and plated for phage titration using the DLA method. On the next day, plaques were counted and PFU/mL was calculated as mentioned above.
3.11. Isolation of Bacteriophage Genome
The extraction of phage genomic DNA was done according to the Martha and Clokie method with slight modification (26). Briefly, 10 mL of phage lysates (with 10% PEG 8000, and 1 M concentration of NaCl) were incubated overnight at 4°C and centrifuged at 10,000 × g for 15 min. The pellets were re-suspended in 500 ml of SM buffer and transferred to a 2-mL Eppendorf tube. A mixture of 5 µL of 1 mg/ml DNase I and 2 µL of 12.5 mg/mL RNase A were added to the tube and incubated for 30 min at 37°C. Afterward, 12 µL of 20% SDS and 5 µL of 10 mg/mL proteinase K were added to the mixture and incubated for 30 min at 37°C. Extraction was carried out with 0.5 mL of phenol: chloroform: isoamyl alcohol (25:24:1) solution. The mixture was spun for 5 min at 15,000 × g for phase separation. The supernatant was transferred into a fresh tube and extracted once with 0.5 ml of chloroform: isoamyl alcohol (24:1) solution, followed by centrifugation for 5 min at 8,000 × g. The supernatant was transferred into a fresh 2-mL Eppendorf tube and 500 µL of 100% isopropanol and 45 µL of 3 M sodium acetate (pH = 5.2) were added. The DNA was left for 30 min to precipitate at room temperature. Following centrifugation for 20 min at 14,000 × g, the DNA pellet was washed twice with 70% ethanol and left to dry. The dried DNA pellet was re-suspended in 50 µL of distilled water. The phage genomic DNA concentration and quality were determined using a NanoDrop spectrophotometer (Nanodrop One C, Thermo Fisher Scientific, assembled in USA), following the instructions provided by the manufacturer. Then, 10 µL of genomic DNA was resolved by electrophoresis on the 0.1% agarose gel with DNA molecular weight marker (λ-Hind III digest) and D2000 DNA size marker (Takara).
3.12. Restriction Digestion of Phage DNA
The DNA of the isolated bacteriophage was digested for 5 - 16 h using Rnase A, Dnase I, and six restriction endonucleases (EcoRV, EcoRI, HaeIII, XbaI, SmaI, SacII, and EcoRV) purchased from Thermo scientific (EU), Lithuania. After the enzymatic digestion, restriction fragments were separated by electrophoresis on a 1% agarose gel containing safe stain (SMO Bio-FluoroVue Nucleic Acid Gel Stain (pre-stain)) in TBE buffer (Tris-boric acid-EDTA), at 90 V in a peQ Lab agarose gel electrophoresis system (peQLAB, E0303, Taiwan). Gene ruler 10 Kb DNA ladder (Thermo Fisher scientific) was used as a size marker. The gel was visualized in a gel documentation system (Vilber Lourmat, E-Box CX5.TS, France). Restriction digestions were performed in triplicate.
3.13. Bacteriophage Storage
Phage lysate was filtered and precipitated by adding polyethylene glycol 8,000 (BIO BASIC Canada INC. Cat. #: PB0433) and NaCl (Merck, EMSURE® ACS, ISO, Reag. Ph Eur, Germany) to final concentrations of 10% and 1 M, respectively, followed by incubation at 4°C for 18 h. The lysate was centrifuged at 10,000 × g for 20 min. The phage pellet was re-suspended in SM buffer (100 mM NaCl, 10 mM MgSO4, 10 mM Tris-HCl [pH = 7.5]) with 50% (v/v) glycerol and stored at 80°C for long-term use. For short-term use, prepared stocks were stored at 4°C.
4. Results
4.1. Bacteriophage Isolation
A total of six phages were isolated. Phage MH-1 was chosen for host range determination and continuing the study.
4.2. Host Range Analysis
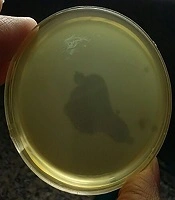
Of the 30 MRSA and 30 MSSA isolates, 27 strains (90%) and 26 strains (86.6%) were sensitive to the isolated phage, respectively, and formed the zone of lysis in the spot test (Figure 1). The lytic activity of the isolated phage was also examined on three other bacterial species. All of the S. epidermidis, E. faecalis, and E. faecium strains were found to be resistant to our isolated phage (Table 1).
| Bacterial Strain | Number of Isolates | Plaque Formation | Reference |
|---|---|---|---|
| MRSA | 30 | 27 out of 30 | Clinical isolates (burn patients) |
| MSSA | 30 | 26 out of 30 | Clinical isolates (burn patients) |
| S. epidermidis | 30 | Not susceptible to phage MH-1 | Clinical isolates |
| E. faecalis | 6 | Not susceptible to phage MH-1 | Clinical isolates |
| E. faecium | 30 | Not susceptible to phage MH-1 | Clinical isolates |
Bacterial Strains Used for Determination of Isolated Phage Host Range
4.3. Morphology of the Lytic Bacteriophage
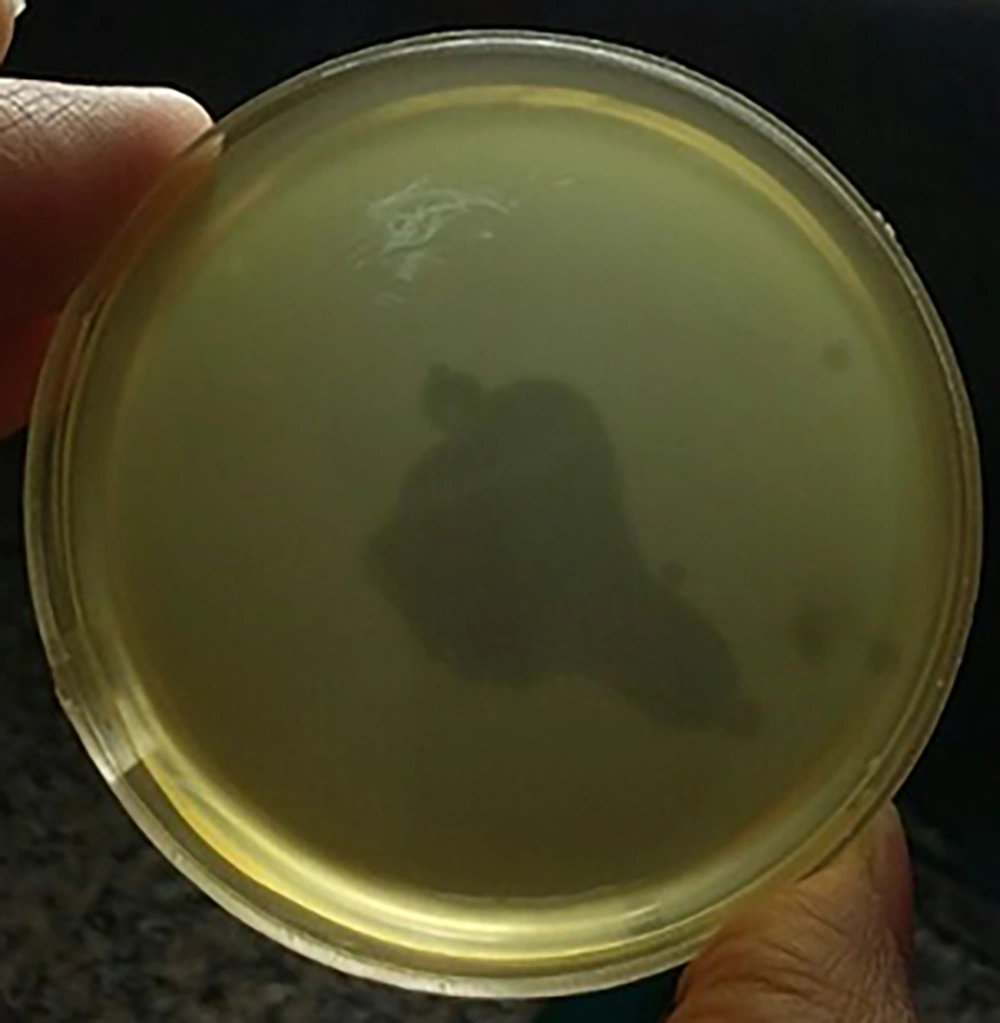
The designation of morphological characteristics by electron microscopy revealed that this phage was a member of the Myoviridea family with a contractile, long, and relatively thick tail and isomeric large head with about 220 and 100 nm in diameter, respectively (Figure 2).
The TEM image of phage MH-1 belonging to the Myoviridea family. A indicates the non-contracted and B contracted tails, respectively. The phages were negatively stained with 1% (wt/v) uranyl acetate and observed using a Zeiss LEO 906 transmission electron microscope (Carl Zeiss LEO EM 906 E, Germany) at an accelerating voltage of 100 kV.
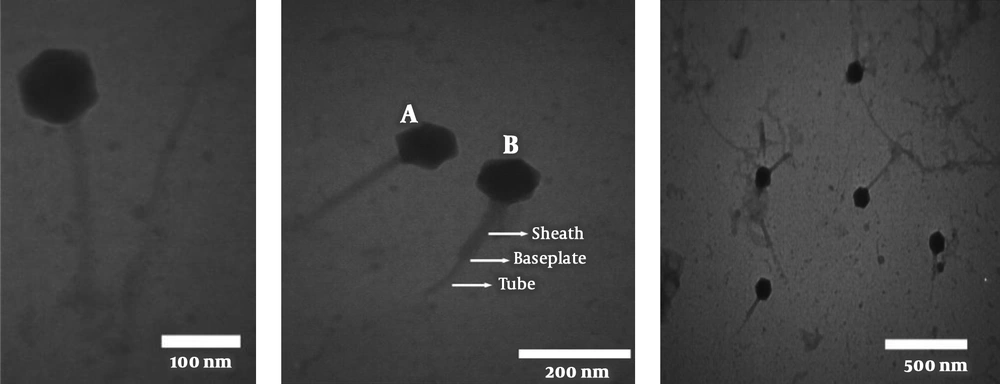
4.4. Determination of Bacteriophage Titer
The phage titer was ascertained by the serial dilution and DLA method. Plaques were counted after overnight incubation at 37°C and the titer of the isolated phage was determined to be 1 × 109 PFU/ml (Figure 3).
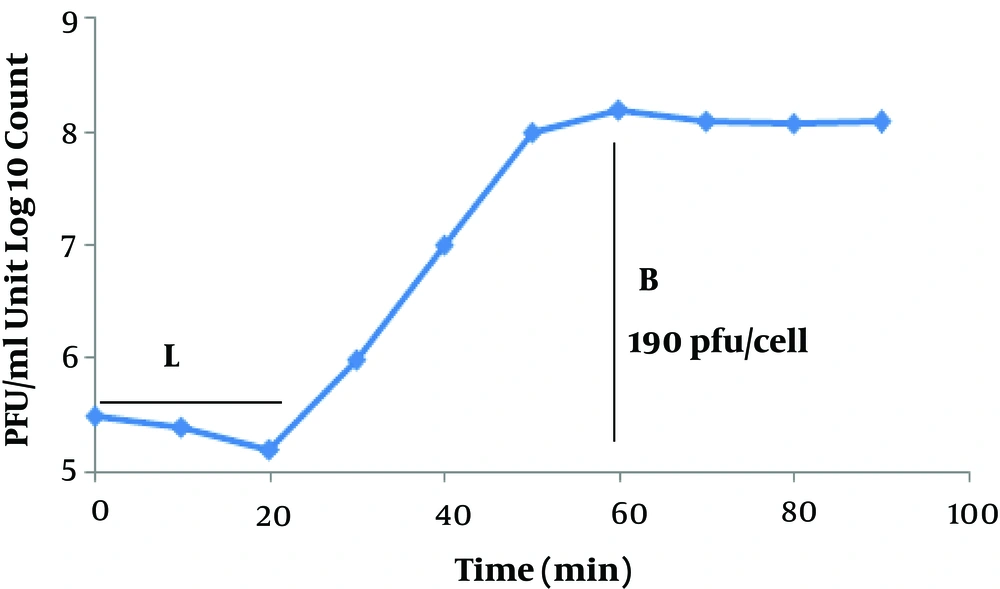
4.5. Latent Time and Phage Burst Size
To determine the latent time and burst size of the phage MH-1, a one-step growth curve analysis was performed. From the analysis of the MH-1 one-step growth curve, the latent period was estimated to be about 20 min. The burst size was 190 PFU per infected cell (Figure 4).
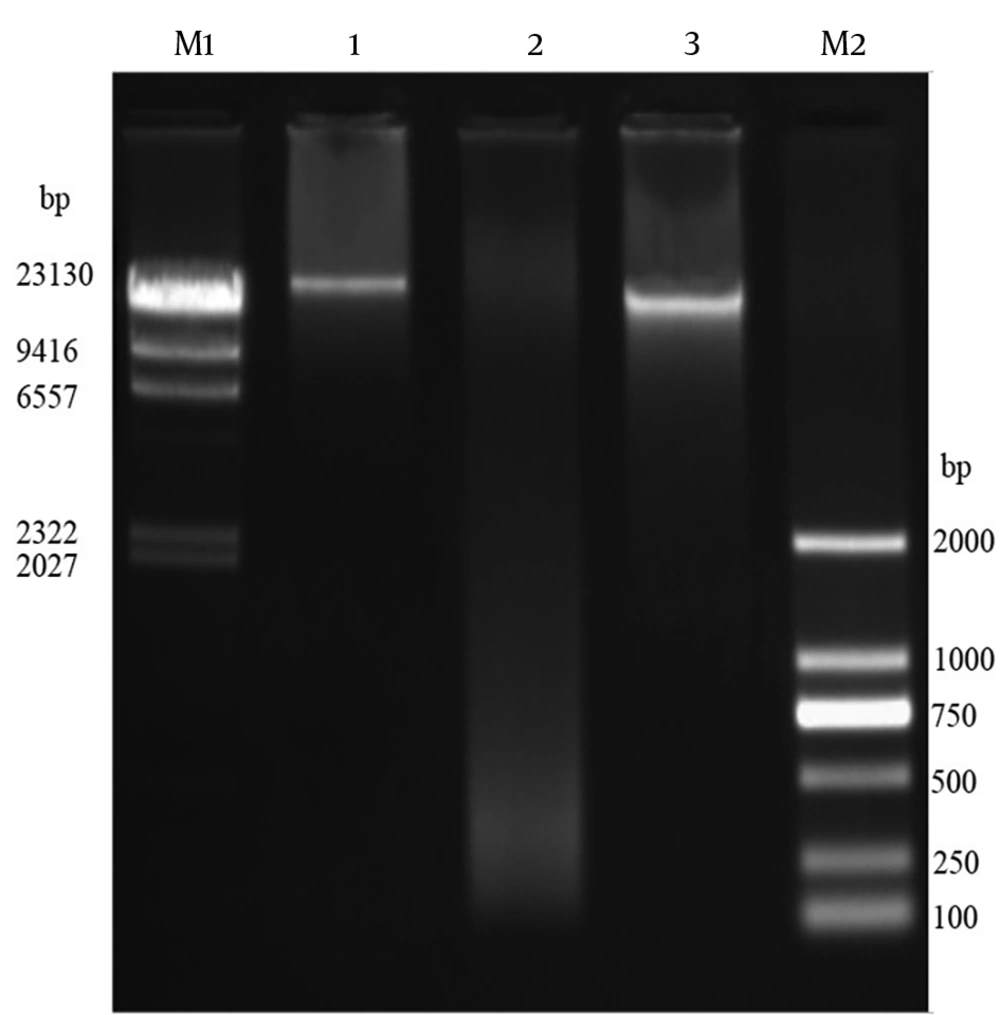
4.6. Isolation of Genomic DNA of Bacteriophages
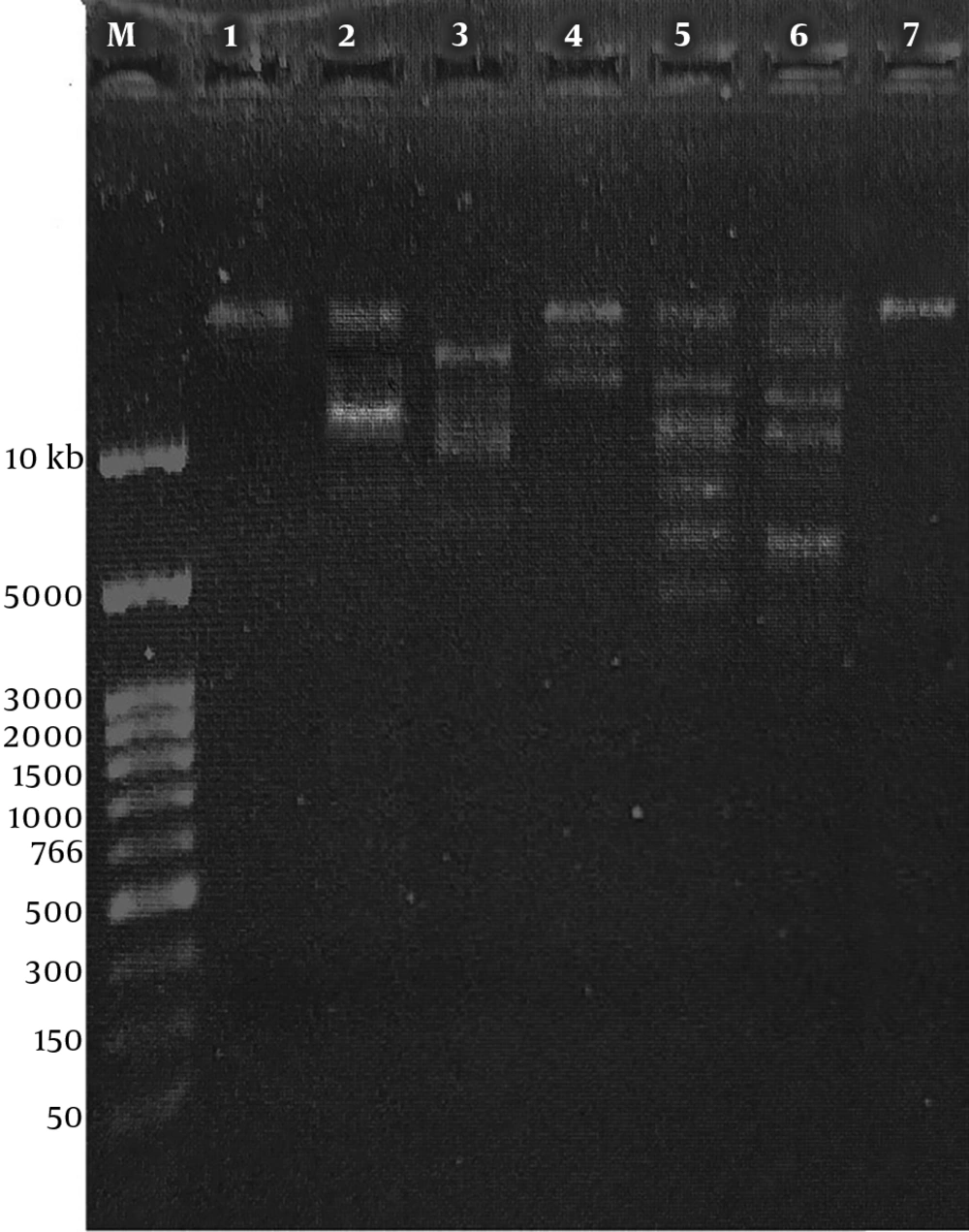
The genomic DNA of phage MH-1 was < ∼23 kbp and it was totally digested by DNase I but not by RNase A (Figure 5). The EcoRI, HaeIII, XbaI, SacII, and EcoRV restriction enzymes digested the phage DNA. However, the phage genome seemed to be resistant to digestion by the SmaI restriction enzyme (Figure 6).
5. Discussion
Treatment of staphylococcal infections is becoming increasingly difficult in view of the widespread presence of MRSA in burn wards (7). Therefore, the necessity for an alternative therapeutic method other than antibiotics seems absolutely crucial. Our study highlights the bacteriophage use as a possible alternative solution. Phage MH-1 showed broad lytic activities (90%) and promising potential for phage therapy.
A few studies are available concerning MRSA bacteriophages in Tehran, Iran, and there is limited information about these local phages and their therapeutic potential. Therefore, in the current study, we carried out a study for the isolation of new staphylococcal bacteriophages that would be specifically active against MRSA strains isolated from burn patients. In the first step, a total of 30 MRSA isolates were collected to be used as hosts. Further characterization was carried out to confirm that the bacteria were S. aureus as expected and to ascertain their resistance to methicillin. Bacteriophage screening, the evaluation of their lytic abilities, and host range determination were the next steps. In the present study, we successfully isolated six virulent phages against MRSA from Motahari hospital sewage over the course of six months. In various studies, such as our study, wastewater has been used as a phage isolation reservoir (29-33). The MH-1 titer was determined to be 1 × 109 PFU/mL; therefore, hospital sewage seems to be a good source for the isolation of this phage. This suggests that potentially valuable therapeutic phages can be easily recovered from such sources and wastewater may have diverse phage populations that could be used for a wide range of applications. Many phages form transparent plaques and are typical of lytic (virulent) phages, whereas phages with the ability to lysogenize host cells (temperate phages) produce opaque plaques. Some phages produce halo plaques, meaning they have semi-transparent areas around the plaques. Halos are due to the release and subsequent activity of soluble enzymes produced by a phage that degrade the cell wall (34). After 18 h incubation at 37°C, the MH-1 phage produced clear plaques without halo with 0.5 - 2 mm in diameter, which is further indicative of the strict lytic nature of the isolated phage.
Phages can detect bacterial cell surface components including lipopolysaccharide (LPS), peptidoglycan, teichoic acids, outer membrane proteins, oligosaccharides, capsules, and type IV fimbria for the binding process. The specificity of the interaction between phage surface structures and host cell surface receptors is more influential on the bacterial host phage range (35-39). In this study, MH-1 showed broad lytic activities on the tested MRSA and MSSA strains collected from burn patients. Most of our S. aureus strains (53 of 60 strains) were lysed by the isolated phage but no plaque production was observed in the examined S. epidermidis, E. faecalis, and E. faecium strains. This result suggests that the isolated phage was completely specific for S. aureus but there was no difference in susceptibility to the isolated phage between methicillin-resistant and methicillin-sensitive strains. Thus, the MH-1 phage can be used as a disinfectant for circulating S. aureus strains in burn care units and prevent the spread of bacteria and antibiotic resistance.
Morphological characteristics of phages can be used for their classification. While there is a variety of different morphological phage types, most S. aureus phages possess icosahedral capsids whit double-stranded (ds) DNA as a genome and belong to the caudovirales order (tailed phages). This order is further classified based on the tail morphology into three major families: Podoviridae (characterized by short non-contractile tails), Myoviridea (equipped with long contractile tails), and Siphoviridae (with long, flexible, non-contractile tails) (40, 41). Electron microscopy observation of phage MH-1 revealed that this phage belonged to the Myoviridea family. Phages from the Myoviridea family have double-layered contractile tails composed of an inner tube covered by an outer sheath and ended by a base plate (Figure 2). The outer sheath contraction of the tail pushes the tube through the bacterial cell wall and creates a channel for the viral genome delivery into the host cell’s cytoplasm (42). There are reports of other studies that have isolated myophages for Staphylococcus aureus (8, 43).
The one-step growth curve was determined to understand the growth of the phage in S. aureus ATCC 43300 as a host. The isolated phage had a short latent period (20 min) and a large burst size (190 PFU/cell). The short latent time showed that the time needed to replicate the virus inside the host is very short and a new generation of phage will be propagated after 20 min. This feature shows a high therapeutic potential for this phage. Moreover, the brut size of this phage could be of relevant interest because it provides high concentrations needed for phage therapy with little propagation. All of these properties make the MH-1 phage a suitable candidate for biocontrol of this resistant bacterium.
The digestion of DNA with DNase I, but not with RNase A, evidenced that the isolated phage had the ds DNA, as expected. To confirm this result, the extracted genome of the phage was digested with six restriction enzymes. The isolated phage DNA samples were sensitive to EcoRV, EcoRI, XbaI, and HaeIII, and exhibited different restriction endonuclease patterns. Additionally, it seems that the genomic DNA of the isolated phage lacked target sequences recognized by the SmaI restriction enzyme. Phages have developed different anti-restriction strategies against restriction-modification systems (R-M) of bacteria. This system operates a defense against phage infections by means of endonuclease and methyltransferase enzymes (44). The R-M system protects methylated bacterial DNA and cuts off the foreign unmethylated DNA in the identical sequence. If oligonucleotide recognition sequences are present, an unmodified DNA molecule will be hydrolyzed by restriction endonucleases. Point mutations or acquisitions of the cognate methylase gene are the strategies used by bacteriophages to change endonuclease recognition sequences in their genomes (44). However, for the exact determination of recognition sequences in the DNA of phage MH-1, complete genome sequencing is required.
In this study, phage MH-1 was isolated and characterized as a new biological strategy to prevent MRSA infection in burn patients. Its specificity, remarkable lytic effect, and broad-host-range for MRSA strains emphasized that it has a considerable potential to use for prophylaxis and treatment of staphylococcal infections. Because of these features, the MH-1 phage can also be used as a component of anti-staphylococcal and other antibacterial cocktails. It is obvious that additional studies are required to identify the lytic effect of this phage on MRSA isolates from different sources. Additionally, in vivo studies on experimentally induced animal or human infections should be carried out to fully ascertain the phage therapeutic potentials.
5.1. Conclusions
The isolation of specific lytic phages to use against multidrug-resistant Staphylococcus aureus, especially in burn patients, can be promising for the treatment of MRSA infections in the future. In addition, given the cost imposed by mortality and morbidity due to the widespread presence of multidrug-resistant S. aureus strains and the lack of an effective solution to this problem, the evaluation of phage therapeutics in human disease management seems to be a reasonable proposition. As a result, phage therapy can be an alternative to antibiotics to replace them when they fail. This treatment may help prevent potentially fatal infections in the hospital setting. Moreover, the substantially lower cost of phage therapy is another important reason for its broader consideration in the current period of the global crisis in antibiotic resistance and health care economy.