1. Background
Vaginal infections are common problems among women, with more than 10 million annual cases in the United States (1). Although the disease is rarely life-threatening, it is associated with high treatment costs, infertility (1), premature labor, and pelvic inflammatory disease (2). Bacterial vaginosis (BV) is the most prevalent vaginal disorder in women of reproductive age (3). It is also the most common cause of abnormal vaginal discharge in women of childbearing age and has been implicated as a cause of the pelvic inflammatory disease (4, 5).
Gardnerella vaginalis, as the main cause of BV, is an anaerobic, catalase, and oxidase-negative bacterium with a mean size of 0.5 to 1.5 mm (6). It is mainly considered as a part of the lower female genital tract microflora (7) and can also be routinely isolated from the male urogenital tract (8-10).
Diagnosis of BV is based on clinical Amsel criteria and direct Gram-stained smear of vaginal secretions. The BVBlue system (Gryphus Diagnostics, L.L.C.) is a chromogenic diagnostic test based on the presence of elevated sialidase enzyme in vaginal fluid samples. The molecular quantification of G. vaginalis (DNA level, ≥ 109 copies/mL) has the highest diagnostic value for the diagnosis of BV, with excellent sensitivity (95%) and specificity (99%) (11).
Although microscopy and culture associated with clinical symptoms are very useful for the diagnosis of bacterial vaginitis, these techniques are not able to differentiate G. vaginalis from other causative agents of vaginal infections.
2. Objectives
In this study, molecular diagnosis of G. vaginalis in vaginal samples of women referring to gynecology clinics in Chaharmahal and Bakhtiari, Iran, in 2017 was investigated.
3. Methods
In this descriptive research, the study population consisted of symptomatic women referring to gynecology clinics in Chaharmahal and Bakhtiari, Iran, in 2017. After obtaining informed consent, a swab from vaginal discharges of 635 women with symptoms of vaginal infections referring to gynecology clinics in Chaharmahal and Bakhtiari in 2017 was collected by a gynecologist. Each swab was then transferred to tubes containing 1 mL sterile saline. A drop of 1 mL of samples was used for microscopic examination. Another drop of each sample was transferred to the Colombia agar medium for culture. The remainder of each 1 mL sample (about 800 µL) was used for the molecular experiment. DNA from clinical samples was extracted using the QIAamp DNA Mini Kit (QIAGEN, USA) in combination with pretreatment with proteinase K and mechanical homogenization. Briefly, vaginal specimens were vortexed vigorously, and 600 mL of the aliquot of UTM-RT transport medium was added and combined with 60 mL proteinase K (Qiagen). GAACCCGTGGAATGGGCC was used as the forward primer and GGGCGGGCTAGAGTGCA as the reverse primer. DNA samples were then amplified using the polymerase chain reaction (PCR) method. Finally, sequences of the products of the PCR were then determined. Species delineation was based on the comparison of the sequences with relevant reference sequences in GenBank, using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast). Nucleotide sequence data generated in this study were deposited in GenBank under accession number “MH898665.1”.
Sequence contigs of the samples were aligned and edited visually in consensus positions, and compared with sequences from GenBank using the Sequencher software v.4.1.4 (Gene Codes Corporation, Ann Arbor, Michigan, USA).
4. Results
Of 635 cases, 200 patients (31.4%) were diagnosed with BV according to the microscopy and culture methods. All samples positive for BV based on the culture method were also positive according to the molecular method. However, 3.9% of the samples that were negative based on the culture method were positive using the molecular method (Table 1).
| Diagnostic Test | Positive Results | Negative Results |
|---|---|---|
| Microscopy | 98 | 537 |
| Culture | 200 | 435 |
| PCR | 224 | 411 |
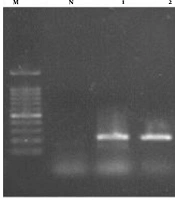
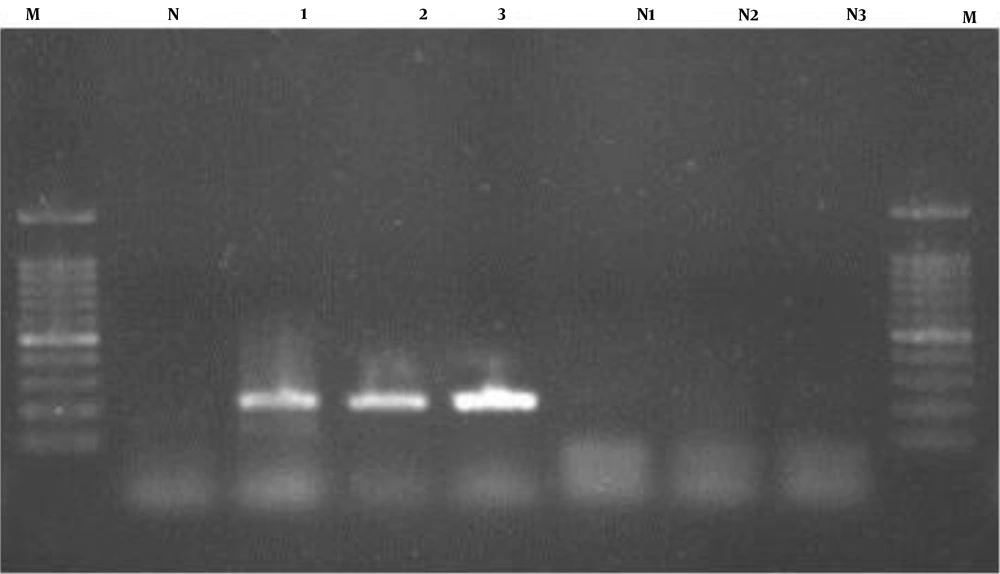
When PCR products of DNA of vaginal samples were subjected to electrophoresis a band of 530bp was detected (Figure 1).
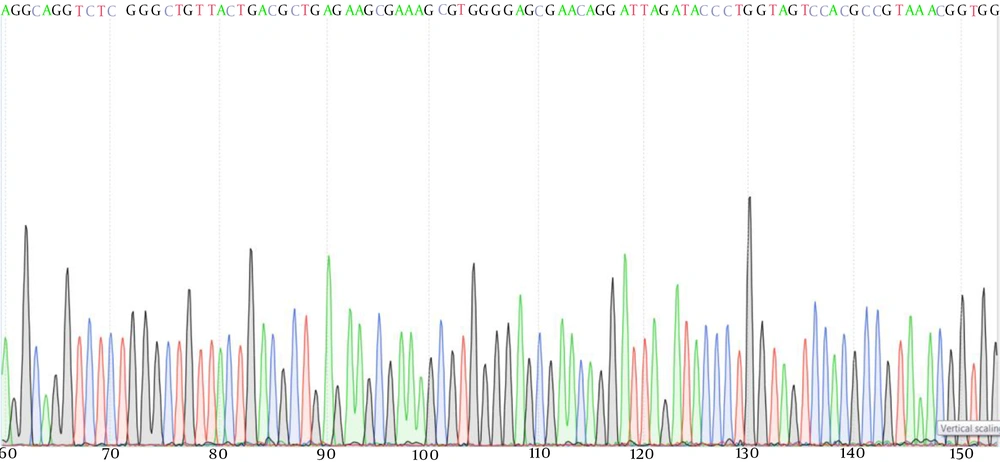
The product of PCR of samples with BV was then subjected to DNA sequencing (Figure 2). After a comparison of the sequences with the reliable sequences in GenBank, the isolated bacteria strain was identified as G. vaginalis.
5. Discussion
Vaginal infections are common diseases of women referring to gynecological clinics all over the world. BV is the most common identified cause of vaginitis and is a common disorder in women throughout the world. Also, G. vaginalis is the most commonly diagnosed agent in the samples collected from cases with vaginal infections. However, no reliable diagnostic tool is available for the detection of this bacterium. Thus, it is necessary to improve diagnostic tests for the diagnosis of this infection, especially for differential diagnosis of BV from other causes of vaginal infections. Molecular methods, such as PCR and real-time PCR are used for the diagnosis of different diseases. However, for developing a simpler method applicable in clinical laboratories, especially in developing countries, we preferred to select a simple PCR method for the diagnosis of G. vaginalis in vaginal samples. In this research, we used a molecular method, in which less equipment and facilities are needed. Our results showed that 31.4% of samples of symptomatic women for vaginal infection were diagnosed with BV. Also, it has been shown that the PCR method is more sensitive than culture for the diagnosis of BV.
Molecular methods have been used in different investigations for the diagnosis of vaginal infections. Cartwright et al. used nucleic acid amplification (NAA)-based assays for the diagnosis of vaginitis in 323 symptomatic women with vaginal infections. They showed that using the NAA-based method resulted in a significant increase in the accuracy of diagnosis of BV in systematic women (12). Sha et al. showed that Amsel criteria had a poor predictive value for the diagnosis of BV. They also reported that the PCR method was significantly more sensitive than Amsel criteria for the diagnosis of this infection (13).
Menard et al. found that molecular quantification of G. vaginalis has 96% sensitivity and 99% specificity (11). Obata-Yasuoka et al. used the PCR method for the diagnosis of BV. They found that based on this method, 20.3% of the vaginal samples were diagnosed with BV. The sensitivity and specificity of multiplex PCR in comparison with Gram staining were 78.4 and 82.9, respectively (14). Lowe et al. used PCR for the diagnosis of vaginal infections. They showed that according to this method, 42% of vaginal samples were diagnosed with BV (15).
Makarova et al. compared morphological, bacteriological, serological, and genetic methods for the diagnosis of G. vaginalis. They showed that PCR was more efficient for the diagnosis of G. vaginalis (16). Menard et al. reported a sensitivity of 95% and specificity of 99% for molecular diagnosis of G. vaginalis (11). Balashov et al. using quantitative PCR identified G. vaginalis infection in vaginal samples (17). Janulaitiene et al. reported that PCR is an efficient method for the detection of G. vaginalis in vaginal specimens (18). Sehgal et al. also reported that qualitative PCR had high sensitivity and specificity for the detection of G. vaginalis and Atopobium vaginae in vaginal samples (19).
The results of these investigations are in agreement with our results, indicating that molecular methods are more sensitive and specific than microscopic methods for the diagnosis of BV. However, they are associated with some limitations to be used in clinical laboratories. Firstly, these methods are time-consuming, and secondly, they need special equipment to be performed. Efforts have been made to develop simple and rapid methods for the diagnosis of vaginal infections (20). Thus, more investigations are needed for finding a rapid and simple method with high sensitivity and specificity.
5.1. Conclusions
Our results showed that PCR is more sensitive than microscopy and culture methods for laboratory diagnosis of BV.