1. Background
Antibiotic resistance is an important warning for infection control and the treatment of infectious disease (1). Multidrug-resistant strains of Pseudomonas aeruginosa (MDRP) have been distinguished to be resistant to carbapenems, fluoroquinolones, and aminoglycosides and are common causative nosocomial infections, especially in immune deficient patients such as those with cystic fibrosis (CF) and burn patients (2, 3). In both burn and CF patients, P. aeruginosa is a major cause of morbidity and mortality among hospitalized patients (4, 5). Most studies on antibiotic resistance mechanisms in P. aeruginosa have investigated modifying enzymes and attaining plasmid resistance genes, the overexpression of efflux pumps, or post-mutational changes in chromosomal and plasmidic genes (6-8). Metallo-beta-lactamases form the most important group of carbapenemases such as blaIMP found in P. aeruginosa. This type is mostly located in class 1 integrons and was discovered in Japan in 1991 (9). GES-1, a plasmid-borne integron detected in P. aeruginosa (formerly, detected among Klebsiella pneumoniae in France in 2000) too (10). The VEB and PER types are the most common ESBLs reported in P. aeruginosa. These types belong to class A of ESBLs and illustrate a high rate of resistance to cephems, monobactams, and ceftazidime (11, 12). The blaVEB-1 gene was first detected in Escherichia coli in a Vietnamese patient and subsequently discovered in P. aeruginosa isolates from a patient from Thailand (10). PER-1 was first discovered in P. aeruginosa isolated from a Turkish patient in France in 1991. The dissemination of PER-1 among European countries with no close geographical vicinity to Turkey, such as Belgium, Italy, and Poland as well as in Asia, has been reported (13). Molecular classification of betalactamas was shown in table 1.PmrA/PmrB proteins as a two component system (TCS), including the response regulator PmrA protein and its consubstantial sensor kinase PmrB, have been distinguished as the main regulatory system in polymyxin B resistance. The activity of this TCS PmrAB in P. aeruginosa has been related to resistance to cationic antimicrobial peptides consisting of colistin (14-16).
The mcr-1 gene was first reported in animal and human isolates of Escherichia coli and Klebsiella pneumoniae in China. An uncommon enzymatic phosphoethanolamine transferase activity (mcr-1 gene product) was detected as a mechanism of colistin resistance related to the enzymes (17).
2. Objectives
Due to increasing of microbial resistance, this study was performed to investigate the frequency of blaIMP, blaGES, blaVEB, blaPER and mcr-1 genes and mutations in pmrA and pmrB genes in P. aeruginosa isolates among CF and burn patients in two educational hospitals in Tehran, Iran.
3. Methods
3.1. Bacterial Identification
This descriptive study was approved by the Research and Ethics Committee of the Science and Research Branch of Islamic Azad University in, Tehran, Iran. According to the physician’s requests, samples collection was done by hospitals laboratories staffs and transferred to the Microbiology laboratory of Shahid Beheshti University of Medical Sciences. From July 2016 to July 2017, 41 isolates of P. aeruginosa from 50 sputum samples of CF patients of Mofid Children’s Hospital (Tehran, Iran) and simultaneously 80 isolates of P. aeruginosa from 250 wound samples of burn patients admitted to the Shahid Motahhari Hospital (Tehran, Iran) were collected. The wound exudates and sputum samples were cultured on blood agar, chocolate agar, and MacConkey agar (Merck, Germany). The quality of the sputum samples was checked by gram stain (18). After 24 hours at 37°C, biochemical tests such as catalase and oxidase tests, triple sugar iron agar (TSI) (Merck, Germany), oxidative fermentation (OF) (Merck, Germany), and other identification tests based on standard diagnostic protocol were performed on grown colonies (18). The identified and confirmed P. aeruginosa isolates were stored in TSB (tryptic soy broth) medium with 20% glycerol at -75°C for further evaluation.
3.2. Antimicrobial Susceptibility Test (AST)
According to Clinical Laboratory Standards Institute (CLSI) guidelines 2016 (19), the Kirby-Bauer disk diffusion method on Mueller Hinton agar (Merck, Germany) was performed. The antimicrobial susceptibility test was applied on ceftazidime 30 µg, piperacillin-tazobactam 100/10 µg, gentamycin 10 µg, cefepime 30 µg, aztreonam 30 µg, imipenem 10 µg, amikacin 30 µg, ciprofloxacin 5 µg, and piperacillin 100 µg. All were purchased from the Rosco Company (Taastrup, Denmark), except piperacillin which was purchased from the Mast Group (Merseyside, UK). P. aeruginosa ATCC27853 was used as a control strain.
3.3. DNA Extraction and Molecular Characterization
DNA was extracted using the boiling method (20). The PCR method was used to detect the presence of the blaIMP, blaPER, blaVEB, blaGES, pmrA, pmrB, and mcr-1 genes. The sequences of PCR primers used in this study are shown in Table 1.
| Primer | Primer Sequence | Product Size, bp | Reference |
|---|---|---|---|
| PER | 340 | (21) | |
| F | 5’-GCAACTGCTGCAATACTCGG-3’ | ||
| R | 5’-ATGTGCGACCACAGTACCAG-3’ | ||
| VEB | 643 | (15) | |
| F | 5’-CGACTTCCATTTCCCGATGC-3’ | ||
| R | 5’-GGACTCTGCAACAAATACGC-3’ | ||
| GES | 864 | (22) | |
| F | 5’-ATGCGCTTCATTCACGCAC-3’ | ||
| R | 5’-CTATTTGTCCGTGCTCAGG-3’ | ||
| IMP | 587 | (4) | |
| F | 5’-GAAGGCGTTTATGTTCATAC-3’ | ||
| R | 5’-GTAAGTTTCAAGAGTGATGC-3’ | ||
| PmrA | 301 | This study | |
| F | 5’-CGACGACTACCTGACCAAG-3’ | ||
| R | 5’-GTGGACGTGGACTTCGATG-3’ | ||
| PmrB | 1211 | This study | |
| F | 5’-CCTACCACCTCTCGCTGAAG-3’ | ||
| R | 5’-GAAGTGCAGTTCGACGATGC-3’ | ||
| Mcr-1 | 309 | (23) | |
| F | 5’-CGGTCAGTCCGTTTGTTC-3’ | ||
| R | 5’-CTTGGTCGGTCTGTAGGG-3’ |
The List and Sequences of Primers Used in This Study
The PCR mixture included the DNA template, forward and reverse primers, and the master mix (Taq 2X Master Mix- Ampliqon Co.). The PCR programs are listed in Table 2. The PCR products were analyzed by electrophoresis on 1% agarose gel at 100 V for 45 minutes in 1X TBE (Tris-HCl Boric Acid EDTA), and the results were checked by UV irradiation in the gel documentation system. The sequencing of the genes showing the accuracy of the PCR products was performed by the MacroGen Company (South Korea). The sequence analysis was done with Chromas 1.45 software and BLAST in the NCBI (National Center for Biotechnology Information) website. Amino acid sequences were detected using the MultAlin (multiple sequence alignment with hierarchical clustering, F. CORPET, 1988, Nucl. Acids Res., 16 (22), 10881-10890) program and compared with PAO1 as the P. aeruginosa standard strain.
| Gene | Initial Denaturation | Denaturation | Annealing | Extension | Final Extension | Cycle | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Temperature, °C | Time | Temperature, °C | Time | Temperature, °C | Time | Temperature, °C | Time | Temperature, °C | Time | ||
| blaPER | 94 | 5’ | 94 | 45” | 59 | 45” | 72 | 45” | 72 | 5’ | 36 |
| blaVEB | 94 | 5’ | 94 | 45” | 57 | 45” | 72 | 45” | 72 | 5’ | 36 |
| blaGES | 94 | 5’ | 94 | 45” | 58 | 45” | 72 | 1’ | 72 | 5’ | 36 |
| blaIMP | 94 | 5’ | 94 | 45” | 51 | 45” | 72 | 45” | 72 | 5’ | 36 |
| pmrA | 94 | 5’ | 94 | 45” | 59 | 45” | 72 | 45” | 72 | 5’ | 36 |
| pmrB | 94 | 5’ | 94 | 30” | 64 | 30” | 72 | 30” | 72 | 5’ | 30 |
| mcr-1 | 94 | 5’ | 94 | 45̋ | 54 | 45” | 72 | 45” | 72 | 5’ | 36 |
DNA Amplification (PCR) Programs
3.4. Statistical Analysis
Chi-square statistical analysis test was used to determine whether or not there is any significant difference in the antibiotic resistance rate between P. aeruginosa isolated from CF and burn wound infections.
4. Results
4.1. Bacterial Identification
All gram-negative bacilli, oxidase positive, TSI(K/K), OF+/-, motility positive, and cetrimide agar growth positive colonies were identified as P. aeruginosa.
4.2. AST Results
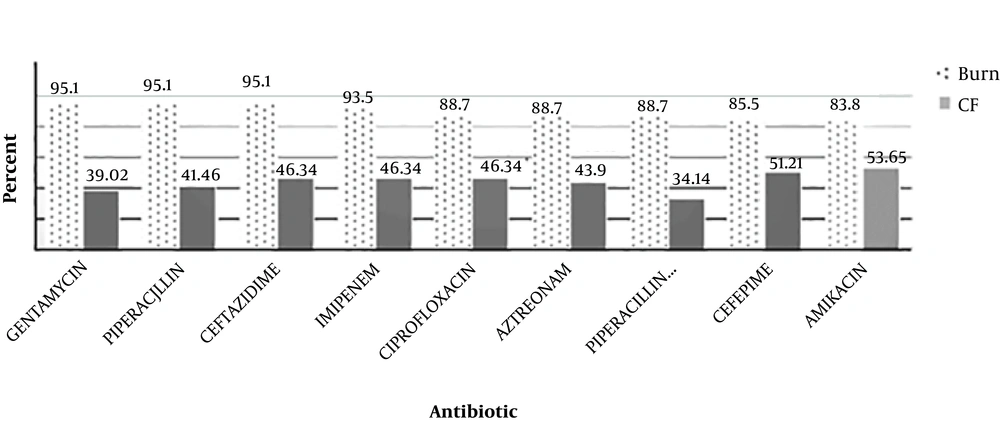
By the AST test, it was shown that among the P. aeruginosa isolates from burn patients, the resistance rate to gentamycin, piperacillin, and ceftazidime was 95.1%, to imipenem was 93.5%, to ciprofloxacin, aztreonam, and piperacillin-tazobactam was 88.7%, to cefepime was 85.5%, and to amikacin was 83.8%. In CF patients, the resistance rate to amikacin was 53.65%, to cefepime was 51.21%, to ciprofloxacin, imipenem, and ceftazidime was 46.34%, to aztreonam was 43.9%, to piperacillin was 41.46%, to gentamycin was 39.02%, to piperacillin-tazobactam was 34.14. The comparison of resistance rates between the two groups of P. aeruginosa isolates is shown in Figure 1. The rate of resistance in burns were higher than CF patients (P ≤ 0.05).
4.3. Molecular Characterization
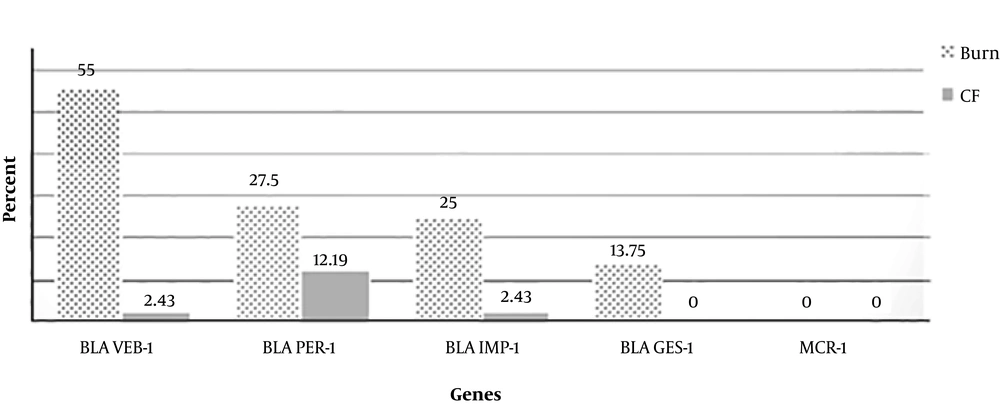
PCR results indicated that among burn patients, 55% of isolates had blaVEB-1, 27.5% had blaPER-1, 25% had blaIMP-1, and 13.75% had blaGES-1. All of the isolates had pmrA and pmrB genes. Interestingly, based on sequencing, no mutation in the pmrA gene was detected in any P. aeruginosa isolates from a CF patient. However, mutations in the pmrB gene in different domains such as (Arg 366 to Gly), (Thr 368 to Gly), and (Pro 369 to Arg) were observed. Mutations detected after the sequence alignment results in pmrA and pmrB genes in burn patients were as follows: (Ala137 to Glu, Gln 146 His, Arg 153 to Pro.) and (Gln 120 Thr, Gly 121 to Ala, Pro 122 to Ser and Leu 137 to Cys).
In CF patients, among 41 isolates of P. aeruginosa, the frequency rates of the pmrA, pmrB, blaPER-1, blaVEB-1, blaIMP-1, and blaGES-1 genes were 100%, 100%, 12.19%, 2.43%, 2.43%, and 0%, respectively.
No mcr-1 gene was detected in any isolate. The comparison of resistant genes between burn and CF isolates is shown in Figure 2. The sequences of blaPER-1 and pmrB were blasted in NCBI and submitted with the accession numbers MF671764 and MF671765 respectively (the remained genes were previously submitted in NCBI). The submission of the pmrA gene and its mutations in NCBI is being processed.
5. Discussion
Recently, P. aeruginosa was identified as the most common pathogen among burn and CF patients who were admitted to hospitals in Tehran, Iran. These opportunistic bacteria can be found in ICU, NICU and other hospital wards. In this study the results of AST showed that the rate of resistance to the tested antibiotics was higher among burn patients than CF patients (Figure 1).
A. Kanayama et al. conducted a study from Jan 2013 to 2014 in Takatsuki, Japan. They determined that P. aeruginosa isolates from different clinical specimens were resistant to imipenem, gentamycin, amikacin, ciprofloxacin, and ceftazidime (minimum inhibitory concentration 4 - 32 µg/mL), and they were susceptible to aztreonam. Similarly, in a recent study, AST was done with similar selected antibiotics based on CLSI 2016. The highest resistance rate was related to gentamycin, ceftazidime, and imipenem in burn patients and to amikacin and cefepime in CF patients (2).
In a 2004 - 2015 study in Latin America, among 3613 P. aeruginosa isolates, the highest susceptibility (72.8%) was seen to amikacin, and 56.8% of the isolates were susceptible to ceftazidime (24). In the current study, however, high levels of resistance were seen to amikacin in burn and CF patients (83.8% and 53.65%, respectively), and it was not a qualified choice for treatment, at least in Iran.
In a study by Hakemi Vala et al. during 2014 in Tehran, Iran, P. aeruginosa isolates from burn patients showed the highest rate of resistance to aztreonam, ceftriaxone, and cefotaxim (82.98%) and to gentamycin, ceftazidime, and piperacillin (95.1%). Such difference is related to the increased use and/or prescription of antibiotics. Also, the other ESBL genes such as blaDIM, SPM, GIM, NDM, VIM, BIC, CTX-M-15 and blaOXA-48 were determined in this study that the frequency of blaIMP was 2.1% (3).
In the 2017 study conducted by Hashem et al. in Ismailia, Egypt, only one (4%) isolate of 147 isolates of P. aeruginosa from different sources had blaIMP, but in the current study, the frequency of this gene was higher among burn patients (25%) (25).
In the study of Kanayama et al. the multiplex PCR results revealed that all isolates had the GES-type β-lactamase gene; in a recent study, however, 13.75% of P. aeruginosa isolates from burn patients had the blaGES gene, but it was not detected in any of the P. aeruginosa isolates from CF patients (2). These differences can be caused by variations in geographical area or the diversity in both countries’ health systems.
Akhi et al. conducted an experiment from July 2008 - 2009 in Tabriz, Iran. Among 56 isolates investigated in their study, 27.5% had the blaPER-1 gene. In the current study, 27.5% (burn patients) and 12.19% (CF patients) of P. aeruginosa isolates contained the blaPER-1 gene. Despite the difference in time, the frequency of this gene has not changed among P. aeruginosa isolates from burn patients. However, CF patients are usually young, and the bacterial isolates from such patients had less resistance because of less antibiotic contact during the patient’s life (26). Thus, a low antibiotic resistance rate among CF patients is predictable.
In their 2012 - 2013 study in Zahedan, Iran, Bokaeian et al. indicated that among 116 P. aeruginosa isolates from different specimens, 13.3% had blaVEB-1. In the current study, however, 55% and 2.43% of P. aeruginosa isolates from burn and CF patients, respectively, showed this gene (27). The reason for the diversity in the results may be explained by not only the difference in the time frame in which these studies were performed, but also by the wide range of patients who referred to Motahari Hospital in Tehran as a reference burn hospital compared to those who referred to the local hospital in Zahedan.
In the study by Moskowitz et al., in 3 alleles, only in a single base (equal to 1-bp) as a transition or transverse mutation was detected. In addition, 6 alleles showed double mutations in 2 missense mutations in the same PmrB molecule. Deletion of a 3-bp fragment including Asp was deleted in locus 45 of pmrB gene. In accordance with the recent study by Moskowitz et al., different mutations were detected in the pmrB gene. However, the similarity between the mutated loci was not confirmed. Moreover, no mutation in the PmrA gene was detected in the recent study among CF patients similar with the Moskowitz et al.’s study (15).
In a 2016 study by Thi Khanh Nhu et al. published in Nature Journal, the PmrAB two component system is recommended as a first genetic mechanism of colistin resistance, pursuant to the results of the current study and other experiments, different mutations can occur in the pmrA and pmrB genes which cause resistance to colistin (28).
In the 2009 study by Barrow and Kwon, after comparing resistant isolates with the PAO1 standard strain, the results revealed that nucleotide substitutions in the PmrB gene were related to two amino acid changes (Ala 247 to Thr and Tyr 345 to His) for the sequence in one of their clinical isolates which showed polymyxin B resistance. They demonstrated that polymyxin B resistance is generally caused by mutations in PhoQ or PmrB (29). In the current study, the pmrB gene showed different amino acid changes as follows: (Ala 1000 to Gly, Gly 1098 to Ala, Ala 1230 to Gly, and Cys 1341 to Gly) in burn patients and (Arg 366 to Gly, Thr 368 to Gly, and Pro 369 to Arg) in CF patients. Also, in a 2012 case report by Lee et al. in P. aeruginosa isolates with a urine source, 3 amino acid substitutions were identified in the pmrB gene (Ala 247 to Thr, Met 292 to Thr, and Tyr 345 to His) (30).
Such variations between the results of the above-mentioned studies and the recent study may be due to differences in the type of antibiotics prescribed, the source of bacterial isolation, or the time difference among their experiment and that of the recent study.
Based on the AST results in the current study, all the P. aeruginosa isolates in burn patients were susceptible to colistin (polymyxin E), but after molecular tests and sequence analysis, the results showed different mutations which represented colistin resistance in these isolates.
The mcr-1 gene as a plasmid mediated gene has been detected mostly in Escherichia coli, but has also been found in Salmonella spp. and Klebsiella pneumonia (31). According to the 2018 study by MacNair et al. which was published in Nature Journal, the mcr-1 gene causes resistance to colistin (32). To the best of the authors’ knowledge, this study is the first of its kind to examine P. aeruginosa isolates for the mcr-1 gene. However, all of the P. aeruginosa isolates were negative for the mcr-1 gene, which indicates that this gene is not common in P. aeruginosa isolates in this geographic region.
Based on Thi Khanh Nhu’s paper, different mutations in pmrAB genes are related to colistin resistance in bacterial isolates (28). Hopefully, by the low frequency of mcr-1 gene and the rate of mutation in pmrAB genes in this study, the rate of resistance to colistin is low here. In conclusion, it can keep low with caution prescription to polymixins.
These finding help to physicians that mcr-1 and pmrAB mutations are not common in P. aeruginosa which indicates that resistance to colistin has not been established yet. So they can control it by correct drug prescription. It is recommended that in clinical laboratories after phenotypic diagnoses, molecular tests such as PCR be used to confirm the results. Molecular tests help physicians be more confident in administering the most effective antibiotic. Unfortunately, most of the laboratories and physicians are not familiar with ESBLs and the standard protocols of detection. Also, patients should desist from using antibiotics arbitrarily and complete their treatment courses. Therefore, the management and treatment strategies should be revised. Due to the importance of the MLST (multi locus sequence typing) or PFGE (pulsed field gel electrophoresis) techniques for determining of the clonality of strains, typing of recent isolates is our future plan.