1. Background
Antimicrobial resistance is still a serious problem throughout the world. Infections caused by resistant organisms are a challenge that requires treatment of both common and life-threatening infections (1). Antibiotics have been widely used today. Cephalosporin used in treating infectious agents has been followed by the emergence and dispersion of extended-spectrum β-lactamases (ESBLs)-producing bacteria that provide resistance to third-generation cephalosporins. Several Enterobacteriaceae in communities in the world have been identified as gram-negative bacteria that produce ESBL (2).
Extended-spectrum β-lactamases are enzymes produced by gram-negative bacteria that can hydrolyze penicillin; first-, second-, and third-generation cephalosporins; and monobactams. ESBL is produced by Enterobacteriaceae, especially Escherichia coli and Klebsiella pneumoniae, which are often associated with healthcare-related infections but are often isolated from bacterial infections that are acquired by the community (3). Particularly, Enterobacteriaceae that are resistant to third-generation cephalosporins have become a major threat now in the communities, not just in hospitals. Enterobacteriaceae is mostly sourced from the digestive tract. Developing countries encounter limited resources and poor hygiene, and this is known to be one of the main causes of the spread of multiresistant bacteria when human excreta reach the environment directly (4, 5).
The simple method with a double-disc synergy test is often used as a screening for ESBL-producing Enterobacteriaceae. The susceptibility of isolates to antibiotics was determined using the Vitek system (bioM_erieux, Marcy l ‘Etoile, France). Then for further analysis, it is necessary to check with the polymerase chain reaction (PCR), which aims to detect ESBL genes. So PCR amplification of bla genes (blaTEM, blaSHV, blaOXA, and blaCTX-M) was carried out using the primers and methods described previously (6).
In general, ESBL-producing bacteria are known to have 3 groups, namely TEM, SHV, and CTX-M types, and Gram-negative β-lactamases are often mediated by blaSHV, blaTEM, and blaCTX-M genes. The SHV enzymes are often associated with Klebsiella pneumoniae and after the active site of the variable sulfhydryl. The TEM was originally found in E. coli in Greece, but its spread has also been very rapid to other genera of Enterobacteriaceae that have been found, such as Citrobacter freundii and Acinetobacter spp. The latest bacteria found not only in hospitals but also in the community are ESBL-producing Enterobacteriaceae that have strong hydrolytic activity against cefotaxime with the help of CTX-M enzyme (7).
Many countries have reported a high prevalence of fecal carriage of ESBLs-producing Enterobacteriaceae in hospitals to the communities, especially in countries with low to medium incomes (4, 8). Even in several countries in Asia, the prevalence of ESBL-producing Enterobacteriaceae has reached up to 70% at the community level (9). As a result of these infections, in some cases, it requires management with expensive drugs and also inpatient hospital care is longer such that it can increase morbidity and mortality (10, 11). During 2008 in Thailand, the bacteria of Enterobacteriaceae were identified in about 60% of the stools of healthy individuals in the community (12). Research in Indonesia showed that the prevalence of ESBL-producing Enterobacteriaceae and the CTX-M gene was about 94.5% (13).
2. Objectives
Because data on the prevalence of fecal carriage of ESBL-producing Enterobacteriaceae in the communities of South Sulawesi is still limited, this study aimed to investigate the prevalence of ESBL-producing Enterobacteriaceae among school children in South Sulawesi, Indonesia.
3. Methods
3.1. Samples and Setting
This study was conducted from September 2016 to November 2018 at the Department of Microbiology, Faculty of Medicine, Hasanuddin University, Indonesia. All stool samples were collected from elementary school students in Sidrap Regency, Makassar City, and Bone Regency, South Sulawesi, Indonesia. Total samples were 245 stool samples with a volunteer sampling technique. Genotype identification was done using polymerase chain reaction (PCR). Written informed consent was obtained from each participant. The study was approved by the Ethics Committee of Faculty of Medicine, Hasanuddin University, Makassar, Indonesia (ref: 1504/H4.8.4.5.31/PP36- KOMETIK/2016).
3.2. DNA Extraction
The stool samples were thawed prior to DNA extraction using the kit from Geneaid. Twenty µL proteinase K was placed in a 1.5 mL microcentrifuge tube, and 500 µL sample was added to the microcentrifuge tube and then was mixed by pulse-vortexing for 10 seconds. Then the sample was incubated at 60°C for 10 minutes and then was shortly centrifuged to remove drops inside the lid. Then 500 µL of ethanol (96 % - 100%) was added to the sample, and then we mixed the sample using pulse-vortexing for 15 seconds. Subsequently, the sample was centrifuged again. Then, the mixture was carefully moved into GD Column (in a 2 mL collection tube) without wetting the rim and centrifuged at 14,000 - 16,000 rpm for 1 minute, and then we transferred the mixture in a clean GD column 2 mL collection tube (provided). In the following, 400 µL of Buffer AW1 was added to the GD Column and centrifuged at 14,000 - 16,000 rpm for 1 minute. Once again, we changed the spin column, and we added 500 µL Buffer AW2. The mixture then was centrifuged at 14,000 - 16,000 rpm for 1 minute. Then the GD Column was transferred in a new 2 mL collection tube (not provided) and centrifuged at 14,000 - 16,000 rpm for 3 minutes. The GD Column was changed with a clean 1.5 mL microcentrifuge tube (not provided), and 100 µL Elution Buffer was added, then incubated at room temperature (15°C - 25°C) for 3 minutes, and then centrifuged at 14,000 - 16,000 rpm for 1 minute. The elution was ready for DNA amplification.
3.3. Detection of ESBL-Producing Enterobacteriaceae Genes by PCR
PCR was used to detect the presence of ESBLs-producing Enterobacteriaceae genes encoding TEM, SHV, and CTX-M-type β-lactamases in stool DNA samples. The selected primers used were SHV-forward: 5’-GGTTATGCGTTATATTCGCC-3’, and reverse: 5’- TTAGGTTGCCAGTGCT-3’ with amplicon size of 747 bp; TEM-forward: 5’- ATGAGTATTCAACATTTCCG-3’, and reverse: 5’-CTGACAGTTACCAATGCTTA-3’ with amplicon size of 445 bp; CTX-M-forward: 5’- ATGTGCAGYACCAGTAARGT-3’, and reverse: 5’-TGGGTRAARTARCTSACCAGA-3’ with amplicon size of 593 bp (14). The primers were provided from Macrogen. PCR was performed on 5 µL of the extracted DNA sample in 25 µL final reaction mixture. The final reaction mixture contained Go taq® green master mix 12.5 µL (Promega), forward primer and reverse at 10 µM 1 µL each. The amplifications were performed consist of initial denaturation at 94°C for 4 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 61°C for 40 seconds), and elongation at 72°C (30 seconds). In the end, the temperature was held at 72°C for 2 minutes. Cycling was followed by a final extension incubation of 2 minutes at 72°C. The PCR products were visualized by running the end product in 2% agarose gel (Merck) stained with ethidium bromide under ultraviolet light. In this study, previously confirmed positive ESBLs genes (TEM, SHV, CTX-M-type) by PCR were used as a positive control, and distilled water was used as a negative control (15).
3.4. Statistical Analysis
The generated data were analyzed using IBM SPSS Statistics 25.0 to give descriptive statistics, which included frequencies and percentages.
4. Results
A total of 245 children were included in the study. The prevalence of fecal carriage of ESBL-producing Enterobacteriaceae in healthy children was 76.7% (188/245). Genotyping of EBSL-producing Enterobacteriaceae encoding genes using PCR revealed that TEM genes (92.6%) were higher than SHV genes (38,8%) and CTX-M genes (4.3%) (Table 1).
| ESBL-Positive Subjects | ESBL-Negative Subjects | |
|---|---|---|
| ESBL-producing Enterobacteriaceae | 188 (76.7) | 57 (23.3) |
| Distribution of ESBL gene from positive-sampel | ||
| TEM | 174 (92.6) | 14 (7.4) |
| SHV | 73 (38.8) | 115 (61.2) |
| CTX-M | 8 (4.3) | 180 (95.7) |
| Totalb | 255 | 309 |
Distribution of ESBL-Producing Enterobacteriaceae and Genotyping of ESBL Genea
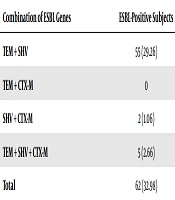
This study found a combined phenomenon of the ESBL-producing Enterobacteriaceae gene. The most combination and the most dominant genes were found TEM + SHV genes of 55 (29.26%) compared with SHV + CTX-M of 2 (1.06%). This study also found five strain carrying the TEM + SHV + CTX-M genes (2.66%) (Table 2).
5. Discussion
The use of antibiotics in Indonesia is still very high, which if its use is not properly controlled, it will later lead to antibiotic resistance, which is a major problem in the health sector today. Feces are a place in which the normal flora of the human intestine grows, especially Enterobacteriaceae. Enterobacteriaceae can change genes because of genetic mutations that will cause extended-spectrum beta-lactamase (ESBL).
Overall, the prevalence of ESBL-producing Enterobacteriaceae in South Sulawesi is very high in healthy communities. Where the ESBL-positive Enterobacteriaceae isolates showed the most encoded TEM gene (92.6%) compared to CTX-M and SHV genes using PCR, this explains that how the spread of epidemic strain clones and the transfer of genetic elements between unrelated strains contribute to the spread of TEM-positive ESBL-producing Enterobacteriaceae in South Sulawesi.
The prevalence of the TEM gene pattern was higher than SHV and CTX-M genes. The results of this study were similar to a study conducted by Livermore (16) on E. coli, who found that the TEM-1 gene was 90%. Similar results were also found in a study in Saudi Arabia, among which 110 positive ESBL-producing isolates of Klebsiella pneumoniae were found in which the TEM gene prevalence was 70.9% (17). Feces may play a role in the spread of Enterobacteriaceae in hospitals and the environment. The prevalence of ESBL-producing bacteria has been reported in Asia. In Indian and Chinese populations, ESBL distribution in the feces may be around 10% (18). Bradford (19) also said that among a number of enzymes associated with ESBL, class A ESBL (including TEM) is the most detected.
However, the results of this study are not in line with the results of research conducted by Tumbarello et al. (20) in 2008 at the Roman Catholic University Hospital, Italy who found that out of 129 different samples, 167 different ESBL-producing genes were evaluated in E. coli in which the most genes obtained were the CTX-M gene, which was 36.5% (61/167), followed by the SHV gene 34.7% (58/167), and the lowest was TEM gene 28.7% (48/167). These differences can be caused by a variety of risk factors, including the geographical location of different regions, the lifestyle between the two populations, and the types of food consumed (20).
This study found a combined phenomenon of the ESBL-producing Enterobacteriaceae gene. The most combination and the most dominant genes were as follows: TEM + SHV genes of 55 (29.26%) compared to SHV + CTX-M of 2 (1.06%). In this study, only one strain carrying the TEM + SHV + CTXM genes (2.66%) was identified among school children. The results of a study in India, where 47 of 82 samples (57.3%) of ESBL-producing E. coli studied had two or more ESBL genes (21). Research was conducted by Tumbarello et al. (20) in 2008 at the Roman Catholic University Hospital, Italy who established that 129 of 167 samples (57.3%) of ESBL-producing E. coli evaluated had two or more ESBL genes. Of 112 fecal samples of healthy children, the transmission of ESBL-producing Enterobacteriaceae was characterized by the discovery of three genes in the same sample of 3 samples (2.6%) positive ESBL-producing E. coli (CTX-M-1, TEM-52, and SHV) -12) (22). Conjugative plasmids play an important role in increasing the spread of ESBL through the transfer of resistance genes to other bacteria in the hospital as well as the environment.
No ESBL gene was found in 57 of 245 subjects (23.3%), including the CTX-M, SHV, and TEM genes, but it did not rule out the possibility that ESBL-negative sample had another ESBL gene that was not evaluated in this study since ESBL has several classes and each class has several genes. Likewise, ESBL-positive subjects can also have other ESBL genes that were not investigated in this study.
5.1. Conclusions
We have shown that Indonesia is another developing country situated in the Southeast Asian region, especially South Sulawesi. Indonesia is affected by the spread of bacterial isolates harboring ESBL genes within the clinical and community environment. The presence of detected ESBL gene careers suggests that antibiotic resistance has spread in the community, thereby needing to be of concern since it can be an ESBL gene reservoir that can be transmitted to many pathogenic bacteria.