1. Background
Similar to eukaryotic apoptosis, bacterial cell death can occur due to the overexpression of some bacterial genes, including toxin-antitoxin (TA) systems or addiction modules whose activity can also inhibit bacterial cell growth through affecting cellular processes. These include DNA replication, macromolecular synthesis, cell wall synthesis, membrane disruption, phage infection, and cytoskeletal polymerization (1, 2).
Toxin-antitoxin systems are purportedly predisposed to lateral gene transfer and intragenic recombination. They were initially discovered in genes carried by plasmids, and were subsequently detected in chromosomal genomes where they are involved in the process of stabilizing bacterial chromosomes and conjugative transposons (3, 4).
A stable protein toxin and a labile protein or RNA antitoxin, which are generally encoded on a single operon, constitute TA modules. Based on the basic characteristics and activity of the antitoxins, TA systems are categorized into six types. Among these, type II, IV, V, and VI TA systems have protein antitoxins; whereas, types I and III have sRNA antitoxins (5).
Several functions have been attributed to TA systems. It has been corroborated that TA modules enable bacteria to evade antibiotic effects on account of inducing a dormant phase and “persister” cell formation followed by the overexpression of numerous toxins (6, 7). In fact, in type II TA systems, labile protein antitoxins can be degraded by Lon and Clp proteases after bacterial exposure to environmental stress, enabling toxins to inhibit bacterial growth by either translation or replication. Furthermore, it has been reported that biofilm formation and quorum sensing, along with phage infection, can be affected by these systems (8, 9).
Shigella flexneri harbors a relatively stable genome and a virulence plasmid, which has been obtained during evolution (10). Similar to Vibrio cholera, S. flexneri is long-lasting in water, causing endemics in developing countries with poor water hygiene (11). This microorganism causes bacillary dysentery in humans, which is characterized by the presence of blood in stool due to the bacterial invasion of the mucosa (12). The primary treatment of this infection is rehydration therapy. However, in severe cases, antibiotics such as ciprofloxacin or co-trimoxazole are prescribed. Unfortunately, there have been many reports regarding antibiotic tolerance in S. flexneri (13, 14). Therefore, it is necessary to find novel strategies to combat this infection.
Identification and confirmation of the presence of TA systems in this microorganism could help us develop novel antimicrobial therapy. One simple method could be the development of a pharmaceutical drug which could alter or destroy the structure of TA complexes. Targeting Lon proteases may be another approach as they play a pivotal role in the activation of toxins in TA systems (15).
2. Objectives
To date, few studies have investigated TA loci in S. flexneri and their role in bacterial survival. Therefore, we aimed to identify TA loci and Lon proteases and study their expression levels in order to evaluate their possible role in bacterial survival following infection of the Caco-2 cell line and to evaluate their potency as new antimicrobial targets for infections caused by S. flexneri.
3. Methods
3.1. Bacterial Strain and DNA Extraction
Shigella flexneri standard strain (ATCC 12022) was used in this study. S. flexneri strain was grown on Hectoen Enteric agar (Merck, Darmstadt, Germany), and the plate was incubated at 35°C in an aerobic atmosphere for 24 h. Further, genomic DNA was extracted using the boiling method (16). A loopful of colonies was suspended in 1 mL of phosphated-buffer saline (PBS). After vortexing, the sample was centrifuged at 14000 × g for 2 min. The pellet was then resuspended in 1 mL of nuclease-free sterile distilled water and subjected to boiling for 1 min. Finally, the sample was centrifuged at 12000 × g for 4 min at 4°C, and after the evaluation of DNA quality, integrity, and concentration by Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, De, USA) and agarose gel-electrophoresis, it was stored in a sterile vial at -70°C for future use.
3.2. Detection of Genes Coding for TA loci and lonP by Polymerase Chain Reaction (PCR)
In order to identify putative TA loci in S. flexneri, various databases, including PSI-BLAST, TBLASTN, and rapid automated scan for toxins and antitoxins in bacteria (RASTA-Bacteria) (17), and TADB 2.0 (18) software were used. Finally, specific primers were designed for the detection of genes coding for putative TA loci and Lon protease in S. flexneri ATCC 12022 using Oligo 7 (19) and Primer 3 (20) software (Table 1).
| Genes | Primer Sequence (5’ → 3’) | Product Size, bp | Tm, °C | References |
|---|---|---|---|---|
| GNAT like-domain | F: CTACGCGCATCGCTAAAGAA | 126 | 60 | This study |
| R: GCGTTGTCTTTCCAGCGAAT | ||||
| RHH-like-domain | F: TCAAAACTAGTCCCGACGCC | 129 | 60 | This study |
| R: GGCGACGTTGGTTATCAAGC | ||||
| yeeU | F: ACCGCCTGTATTACCTTGCC | 81 | 60 | This study |
| R: AGGCCTGGTCCAGATGGTAT | ||||
| yeeV | F: TCTGGCAGACACTGCTCATC | 117 | 60 | This study |
| R: ACAGTGAAATGCCTGCCTCA | ||||
| pfam13957 | F: ACGACGACTCGTTTACCTGG | 108 | 60 | This study |
| R: TTCTGCTGAATTGGCGCAAC | ||||
| COG2161 | F: TCTCTTAAGTGCCAGCGCAT | 101 | 60 | This study |
| R: ATCTTGCAGCACTTGGACGA | ||||
| mazF | F: TGTCACCAGCTAAAAGCCGT | 73 | 60 | This study |
| R: TCAATTTTTCATCGGCAACGGT | ||||
| mazE | F: TTAATTTGCGCGCCACAGAG | 122 | 60 | This study |
| R: CCCTTCAGCCCATCCTTCTG | ||||
| 16SrRNA | F: AGGCCCGAAACTGACGATTT | 166 | 60 | This study |
| R: CATGTCGGCAATGGCATCAG | ||||
| lon | F: CAGCCAGTTCGAAGGCTACA | 172 | 60 | This study |
| R: ACGTTCGTTAACGTCGGACA |
To confirm the existence of genes coding for the studied type II TA system and Lon protease, PCR was performed using specific primers. PCR was performed in a DNA thermal cycler (Bio-Rad, USA) in a volume of 25 µL consisting of 1 µL DNA sample, 1 µL of primers (Forward and Reverse with concentration of 5 pm/µL), 12.5 µL of Taq PCR master mix 2× (Fermentas, Lithuania), and 9.5 µL of DNase/RNAse free distilled water (Thermo Fisher Scientific, US).
The PCR program consisted of an initial denaturation step at 94°C for 5 min; 35 cycles of 95°C for 1 min; annealing at 60°C for 1 min; and extension at 72°C for 25 s. No template control (NTC) was used as a negative control. Finally, PCR products were sent for sequencing (Macrogen, South Korea).
3.3. Cell Culture Infection Assay
To assess the expression levels of type II TA genes and Lon proteases, cell culture infection assay was performed on the human Caco-2 cell line. The Caco-2 cells were grown in T75 flasks containing RPMI medium, 20% fetal bovine serum, 1% sodium pyruvate, 1% non-essential amino acids, and were further incubated in 7% CO2 atmosphere at 37°C. The concentration of 2 × 107 CFU/mL of bacteria was inoculated to each well containing Caco-2 cells. Cells were then incubated at 37°C.
3.4. Evaluation of Type II TA loci and LonP Gene Expression Levels Using Real-Time PCR
After washing steps, 8 and 24 h post-infection, the total RNA was extracted (RNA Extraction kit, Roche, Germany) and cDNA was synthesized (cDNA synthesis Kit, GeneAll Biotechnology, Seoul, South Korea). Then, the expression levels of genes coding for type II TA loci and Lon protease were evaluated by real-time PCR in a rotor-gene thermal cycler (Corbett 6000, Australia) using SYBR green method (AccuPower Green Star qPCR Master Mix, Bioneer, Korea). A total volume of 20 µL reaction containing 1 µL of cDNA, 5 µL SYBR Green master mix, 13 µL nuclease-free water, and 0.5 µL of each primer (forward and reverse, with the concentration of 10 pmol/µL) was run as follows: an initial activation step (hold) at 95°C for 10 min, 45 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 15 s. All tests were performed in triplicates. 16S rRNA gene was used as internal control (Table 1) to normalize target gene expression measurements. Real-Time PCR results were analyzed using the 2 (-delta delta C(T)) method.
4. Results
In order to confirm the presence of type II TA genes in S. flexneri ATCC 12022, four TA systems including pfam13956/COG162, GNAT (Gcn5-related N-acetyltransferase)-like/RHH-like, yeeU/yeeV, and mazF/mazE were investigated by PCR method. Using RASTA-Bacteria and TADB 2.0, it was demonstrated that these TA systems all belong to type II TA systems. The function of the studied putative TA modules were then predicted using bioinformatics databases, namely BLASTP, COG, Pfam, UniProt, and HMMER. Pfam13957 is a toxin belonging to the YafO family toxin with the ability to inhibit protein synthesis by its ribosome-dependent mRNA activity. YeeU is a toxin family that is involved in the positive regulation of cytoskeleton organization. The mazEF system has mRNA endonuclease activity, and GNAT is a protein toxin with acetyltransferase activity. Results of the PCR method showed the presence of these TA systems genes along with Lon protease gene in S. flexneri ATCC 12022.
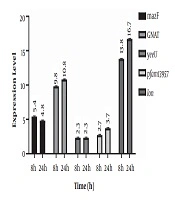
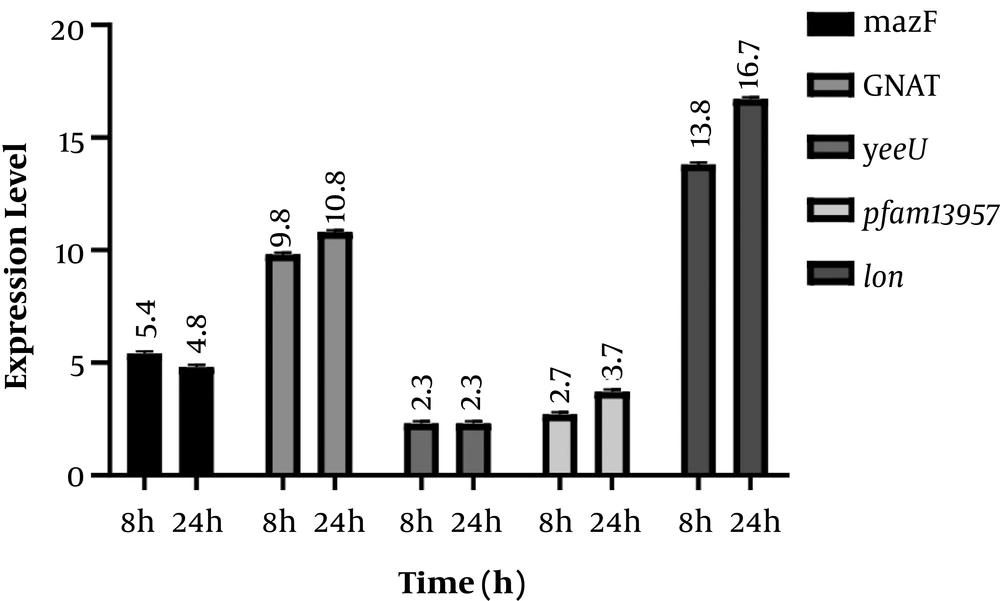
Results of the expression of genes encoding for TA system toxins and Lon protease indicated that after incubation of the infected cell culture, the expression of mazF, GNAT, yeeU, pfam13975, and Lon genes showed 5.4, 9.8, 2.3, 2.7 and 13.8-fold increase, respectively compared to the control. In addition, expression of the aforementioned genes showed 4.8, 10.8, 2.3, 3.7, and 16.8-fold increase 24 h post-infection compared to the control. In fact, GNAT and Lon genes showed significantly higher expression levels compared to the control (P value < 0.05) (Figure 1). However, an increase in the expression level of yeeU was the same at 8h and 24 h post-infection. In addition, mazF expression level showed a slight decrease at 24 h compared to 8 h post infection. Melting curves of the studied genes following real-time PCR are illustrated in Figure 2. Results of the melting curve confirmed the specificity of primers used in this study.
5. Discussion
Several studies have indicated the vital role of toxin-antitoxin systems, including bacterial survival in stress conditions, quorum sensing, and biofilm formation. These systems also account for persister cell formation, making bacteria phenotypically resistant to many antibiotics (21). Furthermore, Lon protease can degrade the antitoxin component of the TA system in stress conditions, facilitating the performance of the toxin component and the subsequent bacterial survival (22). Overall, these systems can be considered a suitable target for finding novel approaches to combat bacterial persistence and infection recurrence. Although many studies have been conducted on TA systems in several pathogenic bacteria (23), only a few have investigated these systems in Shigella flexneri (24, 25). Therefore, the goal of this study was to determine these systems in this bacterium and to investigate their expression following cell infection to determine their contribution to cell invasion and survival. For this purpose, we investigated four TA loci, including pfam13956/COG162, GNATlike/RHH-like, yeeU/yeeV, and mazF/mazE following Caco2 infection. These systems have not been previously reported in S. flexneri in the literature.
Results indicated that 8 and 24 h after the infection of the Caco-2 cell line with S. flexneri, GNAT expression showed 9.8 and 10.8-fold increase compared to the control, respectively (P < 0.05). The GNAT family has been attributed to the acetylation of a variety of substrates, including proteins, small metabolites, and antibiotics such as aminoglycosides (where aminoglycoside N‐acetyltransferases that is involved in antibiotic resistance were the primary GNAT family to be identified) (26). A recent investigation on another GNAT toxin, GmvT, from a virulence plasmid of Shigella sonnei, only demonstrated its role in inhibiting mRNA translation in an acetyl‐CoA‐dependent manner (25). The presence of this system and its significant expression levels following Caco-2 infection in our study may also suggest its role in bacterial survival within cells.
Another studied TA system was pfam13956/COG162. Here, we showed the increased expression of this system for 2.3 and 3.7 fold following cell infection after 8 h and 24 h, respectively. Although the elevation in the expression of these genes was not significant to that of GNAT, however, it may be attributed to S. flexneri survival within cells. Pfam13957 is a putative toxin belonging to the YafO family toxin with the ability to inhibit protein synthesis by its ribosome-dependent mRNA activity. It has been indicated that the overexpression of YafQ can lead to mRNA cleavage preferably at 5’ between the second and third nucleotides in the codon. Although not greatly specific, YafQ has also indicated RNase activity against mRNA, tRNA, and 5S rRNA molecules in vitro (27). Therefore, this system within S. flexneri could also be a suitable target for future research on novel therapeutic agents.
YeeU/YeeV system was another system investigated. YeeU showed the least elevation in expression levels in S. flexneri at both 8 h and 24 h post-infection (2.3 fold increase). This may indicate the least contribution of this system in cell infection and bacterial survival. YeeU-YeeV has been initially considered as one of the main TA systems in Escherichia coli, whose toxin component is involved in blocking the polymerization of bacterial cytoskeletal proteins. YeeU has been indicated to have a specific interaction with MreB and FtsZ, and improve the bundling of their filamentous polymers (28). However, more studies should be conducted to determine the exact role of this system in S. flexneri.
The mazEF system is one of the most well-studied TA systems in bacteria. Here, the expression of mazF showed the second highest increased level among the studied following cell infection with S. flexneri as 5.4 and 4.8 fold increased expression level was observed at 8 h and 24 h post infection. This may also indicate the involvement of this system in bacterial survival. Many studies have shown the contribution of this system in bacterial survival. Sadeghi Kalani et al. showed the importance of this system in L. monocytogenes survival in response as well as biofilm formation (29, 30). Engelberg-Kulka et al. (31) also showed the mRNA endonuclease activity of this system in response to stress conditions leading to the programmed cell death, which can subsequently protect a small population of bacteria in harsh conditions.
Moreover, 8 and 24 h after bacterial invasion of the Caco-2 cell line, the expression of the gene coding for Lon protease showed 13.8 and 16.7-fold increase, respectively, suggesting its importance in bacterial pathogenesis and survival through controlling TA systems. McVicker et al. investigated the role of regulatory proteases, including Lon and ClpP, in the activity of pINV TA systems and showed that Lon and not ClpP is necessary for the activity of TA systems during plasmid stability in this microorganism. In fact, they provided evidence that Lon is the regulatory enzyme for acetyltransferase family TA systems (24).
Similar studies have been conducted on Lon protease in other bacteria. Rogers et al. (32) found that the elimination of gene coding for Lon protease leads to deficiency in biofilm formation and colonization of Vibrio cholera in mice. Niles et al. also investigated the role of Clp protease on type II TA systems in Staphylococcus aureus. They showed that the inhibition of this system has a direct effect on type II TA systems (33). These studies all demonstrate the importance of regulatory proteins Lon and ClpP in TA systems suggesting a possible target for novel antimicrobial therapeutics.
5.1. Conclusions
In general, the current study demonstrated the presence of TA system genes including GNAT-like/RHH-like, pfam13956/COG162, yeeU/yeeV, and mazF/mazE in S. flexneri ATCC 12022. We also showed the elevated expression level of these genes as well as the Lon protease gene following infection of Caco-2 cells, suggesting their possible role in bacterial survival within cells. In fact, genes coding for GNAT and Lon protease showed a significantly higher expression after invading the Caco-2 cell line. Therefore, targeting GNAT or Lon protease can be taken into consideration for future antimicrobial drug evolution. The exact functions and mechanisms of TA systems in S. flexneri isolates are suggested to be further experimentally determined.