1. Background
Staphylococcus aureus, as a Gram-positive round-shaped bacterium is a major human pathogen causing a wide range of nosocomial infections, including wound infections and infective endocarditis. It also results in the development of fatal conditions and life-threatening diseases like bacteremia (1), which if left untreated may lead to the emergence of complications such as systemic inflammatory responses, septic arthritis, and sepsis that, in turn, results in multiple organ damage followed by multiple organ dysfunction syndrome (MOSD). Regarding the management of S. aureus infections in patients with bacteremia, the lack of information on molecular characterization and resistance patterns of S. aureus is taken into account as a major issue (2). This bacterium is known for its unique ability to the acquisition of resistance to antibacterial drugs, especially methicillin. In this regard, methicillin-resistant S. aureus (MRSA) has turned into a major treatment challenge, especially in the Intensive Care Units (ICUs). Methicillin as the first highly-developed therapeutic drug, was administered for the treatment of penicillin-resistant S. aureus infections (3). Resistance to methicillin can be determined according to the expression of mecA gene, which encodes a penicillin-binding protein, known as PBP2a, as a protein with low affinity for β-lactam antibiotics. Moreover, these strains often have a remarkable ability in the acquisition of resistance to a wide range of antibiotics. So far, resistance rates of S. aureus to antibiotics has been a major concern for the treatment of infections. Resistance occurs through drug-inactivating enzymes, modification of antibiotic enzymes, or drug export by efflux pumps. Moreover, S. aureus can encode several multidrug resistance (MDR) efflux pumps (4). Up to now, higher than 10 efflux pumps have been identified, which are encoded by S. aureus. Owing to the presence of fluoroquinolone resistance, extensive use of fluoroquinolone for effective treatment of the infections is restricted (5), which is related to MDR genes, which are chromosomally encoded by norA, norB, and norC, and are widely observed in various isolates and could be specific for a particular substrate or mobilize various antibiotics classes (6, 7). The prevalence of ciprofloxacin resistance in MRSA strains has significantly increased throughout the world, but its resistance in methicillin-susceptible S. aureus (MSSA) has been reported to be low (8, 9).
2. Objectives
In Iran, there is insufficient information about the extent of expression of efflux pump genes in S. aureus strains, which are isolated from blood samples; therefore, this study was conducted to identify antibiotic resistance pattern, and to evaluate the inhibitory effect of efflux pump, MIC of ciprofloxacin, and expression levels of norA, norB, and norC efflux pump genes in the presence of an efflux pump inhibitor against MDR S. aureus.
3. Methods
3.1. Isolation of the Bacteria and Their Characterization
This study was conducted with a cross-sectional design during a complete year (from the first of April 2017 to the end of April 2018). A total of 100 strains of S. aureus were evaluated, which were obtained from the culture of blood samples of the patients referred to different wards in the hospital affiliated with the Shahid Beheshti University of Medical Sciences. Age range of the patients was from 1 to 90 years old. Also, 47% of the patients were female, and the rest of them (53%) were male. Clinical samples were collected using diphasic blood culture media. Suspected positive samples were sub-cultured on a specific medium containing chocolate agar (Merck, Germany) and also MacConkey agar media (Merck, Germany). S. aureus strains were identified by laboratory microbiologic tests like fermentation on salt agar and deoxyribonuclease (DNase) test (Merck, Germany), catalase and rabbit plasma coagulase testing (8).
3.2. Evaluation of the Resistance to Methicillin
Isolates of MRSA were examined using 30 µg cefoxitin disc (MAST DISKSTM, UK) on Mueller-Hinton agar plates supplemented with 4% NaCl, according to the CLSI guidelines (8).
3.3. Antimicrobial Susceptibility Testing
A test was done to assess antimicrobial susceptibility using chloramphenicol (CC 30 µg), linezolid (LZD 30 µg), ciprofloxacin (CIP 5µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), clindamycin (CD 2 µg), ampicillin (AP 10 µg), erythromycin(E 15 µg), and antimicrobial disks using Kirby-Bauer method (MAST DISKSTM, UK) in accordance with CLSI guidelines (8). In addition, S. aureus subsp. ATCC 25923 was applied as a control strain.
3.4. Evaluation of MIC in Staphylococcus aureus Isolates
The Liofilchem® MIC Test Strip (MTS) (Liofilchem Co., Roseto, Italy) was used to specify MIC of S. aureus isolates in accordance with the manufacturer's instructions. The standard reference strain of S. aureus ATCC 25923 was applied as a control strain. MIC breakpoints for vancomycin were determined as follows according to the CLSI guidelines: resistant, ≥ 16 μg/mL; intermediate, 4 ± 8 μg/mL, and susceptible, ≤ 2 μg/mL (8).
3.5. Treatment of Efflux Pump Inhibitor
To find an active efflux pump system, CCCP as an efflux pump inhibitor (from Sigma Aldrich) was supplemented to each M-H agar plate containing 0.5 - 128 μg/mL of ciprofloxacin. The final concentration of carbonyl cyanide 3-chlorophenyl hydrazone (CCCP) in the M-H agar was equal to 25 μg/mL (9). After repetitions several times, MIC was determined for ciprofloxacin with great accuracy (10). A free-antibiotics plate containing carbonyl cyanide m-chlorophenylhydrazone was used as the control. The criterion for positive results regarding the existence of active efflux pump would be a decrease in at least 4 folds of ciprofloxacin MIC after CCCP addition (11).
3.6. Extraction of Genomic DNA
Genomic DNA was extracted using a commercial Kit (Roche, Germany, and Lot. No.10362400) based on the manufacturer’s protocols. Lysostaphin (Sigma-Aldrich, USA) was used at a final concentration of 15 µg/mL for cell wall lysis. Finally, DNA concentration was evaluated using the NanoDrop instrument.
3.7. Technique Used for PCR-Sequencing
Efflux pump genes such as norA and norB as well as norC, gmK, and spA genes were detected using the primers presented in Table 1 (12). According to the procedure proposed by Hu, et al., 2011), PCR was done on a 25-μL mixture containing 1 μL (20 ng) of genomic DNA and 10.5 μL of 2× Master Mix (SinaClon-Iran, CAT. No., PR901638) including 1.5 × PCR buffer, 0.5 mmol/L of dNTPs, 4 mmol/L of MgCl2, and 0.08 IU of Taq DNA polymerase, 1 μL of 10 pmol of each primer and 11.5 μL of sterile distilled water. Amplification was accomplished on a thermal cycler (Eppendorf, Master Cycler Gradient, and Germany). PCR was performed according to the following thermal protocol for each cycle: primary denaturation for 5 min at 95°C, 30 cycles of amplification composed of 30 s at 94°C, annealing at 60°C for 30 s, according to the primers for each gene and at 72°C for 30 s, followed by an extra extension step at 72°C for 5 minutes. Amplified products were electrophoresed by 1 - 1.5% agarose gel. The results were visualized by DNA Safe staining, and images were taken under UV irradiation. The PCR products were purified using a kit (Bioneer Co., Korea), and then, nucleotide sequencing of amplicons was done by an ABI PRISM 3700 sequencer (Macrogen Co., Korea). Sequenced data were analyzed using Chromas software Ver. 1.45 and Nucleotide BLAST program (http://www.ncbi.nlm.nih.gov/nucleotide/).
| Gen | Primer | Primer Sequence (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| norA | norA-F | GACATTTCACCAAGCCATCAA | 102 | (12) |
| norA-R | TGCCATAAATCCACCAATCC | |||
| gmK | gmK-F | TCAGGACCATCTGGAGTAGGTAAAG | 108 | (12) |
| gmK-R | TTCACGCATTTGACGTGTTG | |||
| norB | norB-F | ATGTTTGTCGTTGGAGCAGG | 117 | (12) |
| norB-R | AATACACGCTGCTGATACGC | |||
| norC | norC-F | ATGAATGAAACGTATCGCGG | 130 | In this study |
| norC-R | GTCTGCACCAAAACTTTGTTGTAAA | |||
| spA | spA-F | TAAAGACGATCCTTCAGTGAGC | variable | (12) |
| spA-R | CAGCAGTAGTGCCGTTTGCTT |
3.8. Preparation of RNA and qRT-PCR
Isolates of MRSA were assessed to determine the expression of norA, norB, and norC efflux pump genes. GmK, as an S. aureus housekeeping gene, was used as an internal control. Strains were grown on LB broth overnight (13). Total RNA extraction Kit RNX- Plus (Cat. No., RN7713C, Sinacolon, Iran) was used to extract total RNA according to the manufacturer’s guidelines. Extra DNA was removed from RNA by RNase-free DNase I (Fermentas, USA). Integrity and purity of total RNA were specified using the NanoDrop (WPA Biowave II Nano spectrophotometer, USA). Briefly, cDNA synthesis was performed using the Takara Kit (Japan) (Table 2). Quantitative real-time PCR (qRT-PCR) was performed on synthesized cDNA through the use of the Power SYBR Green PCR Master Mix (Bioneer, Daejeon, Korea) with a Corbett Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science, Australia). Amplification was performed in the following condition: firstly, 10 min denaturation at 94°C, followed by 40 cycles of 20 s at 94°C and 45 s at 59°C. Samples were run in triplicate. Controls were run without the use of reverse transcriptase to confirm the absence of contaminating cDNA in any of the samples. The expression levels of norA, norB, and norC genes were normalized using the gmK housekeeping gene as an internal control, as calculated based on 2−ΔΔCT method. The results were expressed as relative expression of the mRNA with respect to S. aureus ATCC 25923. The Ct parameter (threshold cycle) was defined as the cycle number at which the first detectable fluorescence signal of the reaction crosses the threshold, which began to increase exponentially by binding of SYBR Green I dye to the minor groove of double-stranded DNA.
| Gene | Primer | Primer Sequence (5′-3′) | Purpose | Product Size (bp) | Comment |
|---|---|---|---|---|---|
| norA | norA-F | GACATTTCACCAAGCCATCAA | qRT–PCR | 102 | Target gene |
| norA-R | TGCCATAAATCCACCAATCC | qRT–PCR | |||
| gmK | gmK-F | TCAGGACCATCTGGAGTAGGTAAAG | qRT–PCR | 108 | Internal control |
| gmK-R | TTCACGCATTTGACGTGTTG | qRT–PCR |
3.9. SpA Typing
Molecular epidemiological analysis was performed by spA typing according to the study by Harmsen, et al. (14). When positive spA PCR products were purified using Kit (Roche, Germany), they were tested by DNA sequence analysis and nucleotide sequences were determined on both strands using ABI Prism 377 automated sequencer (Applied Biosystems, Perkin-Elmer Co., Foster City, CA). Chromas software (version 1.45, Australia) was used to edit obtained sequences. Edited sequences were assigned to specific spa types according to the guidelines described by the Ridom spA server database (http://www.spaserver.ridom.de).
3.10. Statistical Analysis
SPSS software, ver. 22.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analyses. A P value of ≤ 0.05 was considered statistically significant.
4. Results
4.1. Patients and Bacterial Isolates
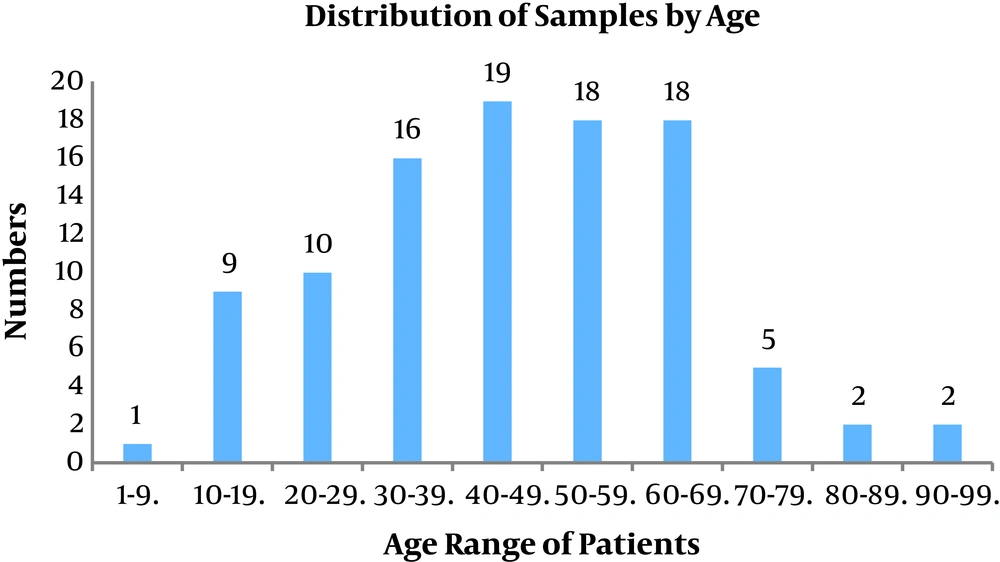
A total of 100 samples of S. aureus isolates were collected from the patients that were referred to different wards in the hospital affiliated to Shahid Beheshti University of Medical Sciences from April 2017 - 2018 of which 53 belonged to males and 47 belonged to females (male:female ratio = 1.12). The patients aged between 1 - 90 years old. All the isolates were prepared from blood samples. Figure 1 presents age of the patients.
4.2. Antibiotic Susceptibility Profile
MRSA isolates were resistant to ampicillin (100%), erythromycin (91.1), cefazolin (82.2%), ciprofloxacin (84.4%), clindamycin (80%), chloramphenicol 8.8%), and TMP-SMX (20%). Interestingly, all MRSA and MSSA isolates showed the highest susceptibility of 100% to linezolid. Table 3 shows the antibiotic susceptibility of the isolates. Among 100 analyzed S. aureus isolates, all isolates were susceptible to vancomycin based on MIC results.
Of ciprofloxacin-resistant isolates, 8 isolates had an active efflux pump, according to CCCP results. Table 4 shows the effect of a pump inhibitor (CCCP) on the treatment of efflux pump.
| Antibiotics | MRSA/(%), N=45 | MSSA/Resistant (%), N=55 | ||||
|---|---|---|---|---|---|---|
| R | I | S | R | I | S | |
| Clindamycin | 36 (80) | 0 | 9 (20) | 7 (12.7) | 1 (1.8) | 47 (85.4) |
| Linezolid | 0 | 0 | 45 (100) | 0 | 0 | 55 (100) |
| Chloramphenicol | 4 (8.8) | 0 | 41 (91.1) | 2 (3.6) | 0 | 53 (96.3) |
| Cefazolin | 37 (82.2) | 8 (17.7) | 2 (3.6) | 0 | 53 (96.3) | |
| Erythromycin | 41 (91.1) | 3 (6.6) | 1 (2.2) | 7 (12.7) | 6 (10.8) | 42 (76.3) |
| Ampicillin | 45 (100) | 0 | 0 | 51 (92.7) | 0 | 4 (7.27) |
| Ciprofloxacin | 38 (84.4) | 0 | 7 (15.5) | 3 (5.54) | 2 (3.6) | 50 (90.9) |
| Trimethoprim/Sulfamethoxazole | 9 (20) | 0 | 36 (80) | 5 (9) | 0 | 50 (90.9) |
Abbreviations: R, resistant; I, intermediate; S, sensitive.
| Isolate Number | MIC Ciprofloxacin | MIC Ciprofloxacin + CCCP |
|---|---|---|
| 62 | 8 | 2 |
| 100 | 16 | 4 |
| 87 | 32 | 4 |
| 38 | 64 | 8 |
| 1 | 64 | 16 |
| 15 | 64 | 16 |
| 31 | 128 | 32 |
| 39 | 128 | 32 |
4.3. Prevalence of Resistance Genes
Staphylococcus aureus was identified by 16SrRNA. In this regard, norA and norB were present in 100% of the isolates, and norC, gmK, and spA were present in 95%, 98%, and 99% of the isolates, respectively.
4.4. Gene Expression Analysis of norA, norB, norC, and spA Typing
To assess the expression of norA, norB, and norC efflux system in MRSA ciprofloxacin-resistant isolates (MIC ≥ 4 μg/mL), qRT-PCR analysis was performed. According to the results, of 100 S. aureus strains, the norA gene was found as the most commonly overexpressed gene that was present in 7 (63.6%) strains. Only 2 strains (18.2%) showed overexpressed norB gene and 2 strains had overexpressed norC gene. The strains with overexpressed norA, norB, and norC genes were ciprofloxacin-resistant to MRSA. Analysis of 41 ciprofloxacin-resistant S. aureus strains showed norA overexpression, which was more common between ciprofloxacin-resistant strains to MRSA than ciprofloxacin-resistant strains to MSSA. Analysis of 11 MRSA strains resistant to ciprofloxacin showed that increased expression of norA, norB, and norC is associated with a decrease in CCCP of the ciprofloxacin MIC. The results revealed four different types of spA, and the most common types were t037 and t790 (23.3%), followed by t030 (14.1%) and t044 (12.2%).
5. Discussion
The emergence and prevalence of antibiotic-resistant bacteria have resulted in the discovery of new antibacterial agents and modulators of antibiotic resistance. Different mechanisms exist for antibiotic resistance in S. aureus. Efflux pumps, as one of the prominent mechanisms that causes extraction of antibiotics and decrease intracellular concentration of antibiotic (13, 15). In the case of S. aureus, the MDR efflux pumps, namely norA, norB, and norC , which are encoded chromosomally, are observed in different strains, and they have been identified based on their ability to acquire resistance to fluoroquinolones (12). The results of the study demonstrated that resistance to MRSA isolates, including ciprofloxacin (84.4%), erythromycin (91.1%), clindamycin (80%), ampicillin (100%), and cefazolin (82.2%) was relatively high, while approximately less than half of the strains showed resistance to chloramphenicol (8.8%) and trimethoprim-sulfamethoxazole (20%), which is in accordance with the findings reported in the study by Ko et al. who investigated 74 MRSA strains isolated from a total of 12 Asian countries (15). To date, due to the significance of the efflux pump in antibiotic resistance mainly in MDR S. aureus, researchers have conducted several studies in this field. For instance, in the present study, the prevalence of ciprofloxacin-resistance was equal to 84.4% in MRSA isolates, while it has been reported between 29 - 99% in other studies (16, 17) conducted in Tehran. All MRSA isolates showed susceptibility to linezolid and vancomycin; hence, yet these antimicrobial drugs can be administered for severe MRSA-induced infections in Iran, as reported in the studies carried out by Valadan Tahbaz et al. (18). In the current study, a high rate of resistance was observed to ciprofloxacin, which is related to the effect of permeability, efflux pump, and the decrease in the availability of quinolones at the target site (19). Frempong-Manso et al. introduced norA as the gene most commonly found in the isolates, which its overexpression resulted in low-to-moderate increases in the MICs of fluoroquinolones that, in turn, led to the emergence of high-level target-based resistance in vitro (20). In another study, norB was identified as the most commonly overexpressed MDR efflux pump gene in the isolates prepared from clinical samples from patients in Korea. In a previous study, overexpression of norA gene has been reported in only 2 strains (3.2%) and overexpression of norC has not been found (12). In this study, overexpression of the norA and norB efflux pump genes increased in the presence of inhibitors. DeMarco et al. stated that among S. aureus isolates collected from blood samples, 25.4% of them showed norB overexpression, followed by 22.8 and 16.7% overexpression in norA and norC genes, respectively. It has been found when an overexpression occurs in a single efflux pump gene, it mainly belongs to norA, whereas overexpression of norB and norC genes occurs mostly when two or more efflux pump genes are overexpressed (21). Similar to the study conducted by Kwak et al., in this study, all clinical isolates with overexpressed efflux pump genes showed overexpression in a single gene. In a prior study, only one isolate (0.9%) showed overexpression in both norA and norB genes. In addition, norC has been identified as the only gene, which no overexpression was detected for it among clinical isolates (12). Kosmidis et al. in a study found that overexpression of MDR efflux pump genes changed temporarily and with respect to geographical locations in the isolates collected from clinical samples. They also revealed the predominance of overexpression only for norB in strains isolated from the patients in San Francisco, USA, to the extent greater than any other location (22). Further studies are needed to determine norA, norB, and norC genes overexpression in order to see whether severe effects have occurred on biocide and fluoroquinolone resistance in S. aureus strains (22, 23). Inhibitors causing resistance to bacteria tend to exhibit a likelihood for the treatment of the patients suffered from antibiotic-resistant infections. Applying inhibitors may provide the chance of another treatment for the patients undergone treatment with ineffective antibiotics in health centers and also protects them against new MDR strains (23). Khan observed that the presence of active efflux pumps in all S. aureus isolates is associated with an increase in the ethidium bromide uptake and a decrease in the antibiotic MICs when there are efflux pump inhibitors, e.g. CCCP and reserpine (24). In the present study, among ciprofloxacin-resistant isolates, 11 samples had active efflux pumps according to CCCP results. According to the spA typing results, four different types of spA were determined among 45 MRSA isolates. Although the distribution of spA types in S. aureus strains was different with respect to geographical location. The findings of the studies conducted in Iran demonstrated great dissemination of spA types of t790, t030, t037, and t044 in S. aureus clinical isolates (25-27). The t037 and t790 were the first commonly identified spA types in the current study, accounting for more than 50% of all isolates. These spA types have been reported as predominant spA types in the studies conducted by Goudarzi et al. (28, 29). SpA types of t037 and t790 were found as resistant to methicillin. The t030 was the second commonly identified spA type in the current study, which was found in a single strain. In contrast with prior studies conducted in Iran (30, 31), in this study, a low frequency of t030 and t044 spA types was also found between the isolates.
5.1. Conclusion
Findings of the current study showed significant roles of norA, norB, and norC efflux pump genes in the acquisition of resistance to ciprofloxacin in the S. aureus isolates collected from clinical samples. Seemingly, it is necessary to assess resistance and virulence of genes in different molecular types of S. aureus in order to administer proper antibiotic agents.