1. Background
Regular exercise has beneficial effect on cardiovascular health and function via reducing morbidity and mortality related to cardiovascular disease. This protective effect is mostly based on the metabolic alterations that affect cardiovascular risk factors, including hypertension, low-density lipoprotein cholesterol (LDL-C) and non-HDL-C, body weight and type 2 diabetes mellitus (1, 2). It is also well-documented that most cases of sudden death are related to exercise and are accepted to have a cardiovascular origin in most cases (3). However, molecular mechanisms leading to cardioprotection are not clear (4).
Exercise is a type of regular physical activity within a period of time and intensity, which keep the body fit and healthy and improve and/or maintain physical performance (5, 6). Exercise is recommended as the preventive, curative, and rehabilitative programs to improve and maintain health; especially of the heart (6). The European Society of Cardiology in 2016 released a guideline which recommended aerobic exercise as a model to improve cardiovascular endurance. Aerobic intensity has been divided into 30 minutes/day, 5 days/week of moderate intensity (150 minutes/week), or 15 minutes/day, 5 days/week of vigorous intensity (75 minutes/week), or a combination of both performed in sessions with a duration of at least 10 minutes. Eventhough exercise has been extensively studied, molecular physiological and biochemical mechanisms involved in mediating exercise-induced cardiovascular effects remain unclear (1).
Swimming as aerobic exercise increases oxygen demand which is required in muscle contraction. This condition induces imbalance of the level of oxygen demand and supply in cells which is called hypoxia. This condition is known to occur in skeletal muscle tissue, but the heart muscle as an involuntary muscle that always contracts plays a special role. In the face of various loads, continuous availability of oxygen and other substrates is necessary to preserve it (7). Physiologically, ischaemia, an insufficient blood flow to provide adequate oxygen that leads to tissue hypoxia alters the expression of peroxisome proliferator activated receptor-gamma coactivator 1α (PGC-1α) as a potent sensor of metabolic changes and regulator in alterating the body’s metabolism and hypoxia-inducible factor (HIF-1α) as the master regulator gene expression of angiogenesis (8, 9).
Although both factors play a role in conserving energy during metabolic processes, its role in molecular adaptation to long-term moderate intensity such as swimming in the myocardium is still unclear. Therefore, we investigate the effect of swimming exercise on PGC-1α and HIF-1α gene expression of cardiac muscle in mice.
2. Methods
2.1. Animal
Eight weeks old male BALB/c mice (Mus musculus) were obtained from the Animal Centre of Universitas Padjadjaran. The environment was maintained in a dark light cycle- (12-h light cycle and 12 hours dark cycle) and temperature (22°C). Mice were fed a pellet rodent diet ad libitum and had free access to water. After one week acclimatization period, 14 male mice were divided into two groups, control group and the exercise group.
All experimental procedures followed guide for the care and use of laboratory animals (10) and were approved by Health Research Ethics Committee Faculty of Medicine Universitas Padjadjaran No 706/UN6.C.10/PN/2017.
2.2. Swimming Apparatus and Training Protocol
We designed a swimming apparatus with the size of the outer tank being: 30 cm in diameter, 75 cm in width and 40 cm in height, then the inner tank was divided into 10 lanes with a surface area of 15 × 15 cm per lane and a depth of 35 cm to allow individual training. To prevent floating during the swimming session, water bubbling was produced by tubes connected to an air pump system. A heating system kept the water temperature between 30°C and 32°C.
2.3. Measurement of PGC-1α and HIF-1α mRNA Expression
After four weeks, control and exercise group were anesthetized with isoflurane and followed by cervical dislocation technique, the hearts were removed. The hearts were kept in Eppendorf tube, snap-frozen in liquid-nitrogen and stored at -80°C until RNA isolation.
Total RNA was extracted and isolated from heart using TRIzol reagent (Thermo Fisher Scientific, Uppsala, Sweden). Total RNA in heart were quantified using Multimode Microplate Reader at 268/280 nm absorbance spectrophotometry (M200 Pro, Tecan, Morrisville, NC). Semi-quantitative PCR was performed using the One Step RT PCR Kit (Qiagen, Valencia, CA) and Sensoquest Genecraft. Semi quantitative gene expression levels were normalized using mouse β-actin. Primers sequences used for mouse gene is listed in Table 1. The band of PCR products were visualized with and quantified using ImageJ software (NIH).
| No | Name Gene | Primer | Sequences |
|---|---|---|---|
| 1 | PGC-1α | Forward | 5'-CGCACAACTCAGCAAGTCCTC-3' |
| Reverse | 5'-CCTTGCTGGCCTCCAAAGTCTC-3' | ||
| 2 | HIF-1α | Forward | 5'-CTCCCATACAAGGCAGCAGAAA-3' |
| Reverse | 5'-CAAAACAACCAACAGAAACGAAAC-3' | ||
| 3 | β-actin | Forward | 5'-TGGAGAAGATTTGGCACCA-3' |
| Reverse | 3'-CCAGAGGCATACAGGGACAA-3' |
Primer Sequences
2.4. Hematoxylin and Eosin Staining
Mice were perfused with phosphate buffered saline under deep anaesthesia using sodium pentobarbital. Hearts were taken and incubated with 4% Paraformaldehyde in 4°C overnight. The heart samples were subjected to paraffin embedded blocks and sliced in 50 μm thicknesses. The sections were mounted in a slide. All slides were stained with Hematoxylin-Eosin and Hiedenhain’s Azan staining (Thermo Scientific Fisher, Uppsala, Sweden). The images of heart section were taken using Zeiss Imager. Z2, ApoTome.2 (Carl Zeiss, Germany). Images were processed using ZEN software (Carl Zeiss, Germany)
2.5. Statistics
All statisics were computed using SPSS 20.0 software. Results are presented as the mean ± standard error of mean (mean ± SEM). One Way ANOVA and Tukey post hoc test were conducted to examine mean differences between the groups with 95% confidence interval (P < 0.05)
3. Results
3.1. Body Weight and Heart Weight in Swimming
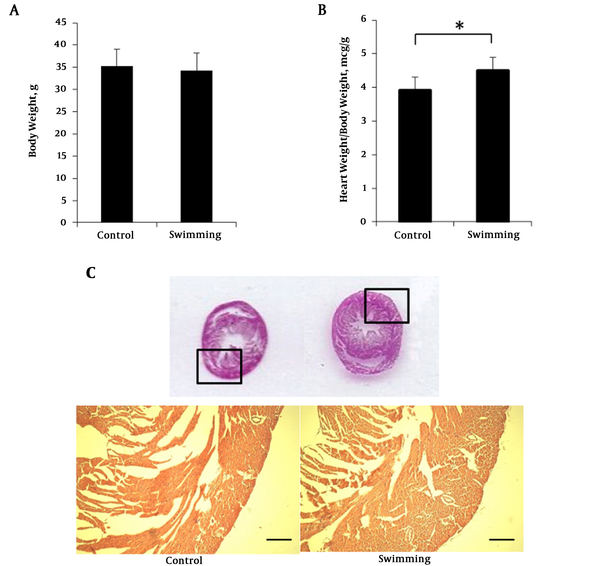
Before the training period, mice were habituated for one week with the swimming chamber. To investigate the body weight change, mice body weight were measured after a training period. Body weight of swimming group (34.32 ± 3.91 g) were not significantly different with the control group (35.207 ± 3.86 g) after the 4 week experimental period (Figure 1A). Interestingly, heart weights were significantly higher in the swimming group (4.51 ± 0.522 g, P < 0.05, n = 7) compared to the control group (3.92 ± 0.377 g, P < 0.05, n = 7) (Figure 1B). Although heart size was almost similar (Figure 1C), the myocardium of the swimming group was thicker compared with the control. The hematoxylin eosin staining from paraffin embedded slice is presented in Figure 1C. All the results convey that 4 weeks swimming training did not change the body weight but increased heart weight due to aerobic adaptation. Increase of heart weight is caused by heart muscular hypertrophy (Figure 1C).
Body weight, heart weight and HE staining swimming mice. A, mice body weight at the end of swimimng period; B, ratio heart weight normalized with body eight; C, hermatoxylin eosin staining of mice heart paraffin section. One way ANOVA test was performed for A and B data presented as mean ± SEM. *P < 0.05.
3.2. HIF1-α and PGC-1α mRNA Expression in Mice Heart
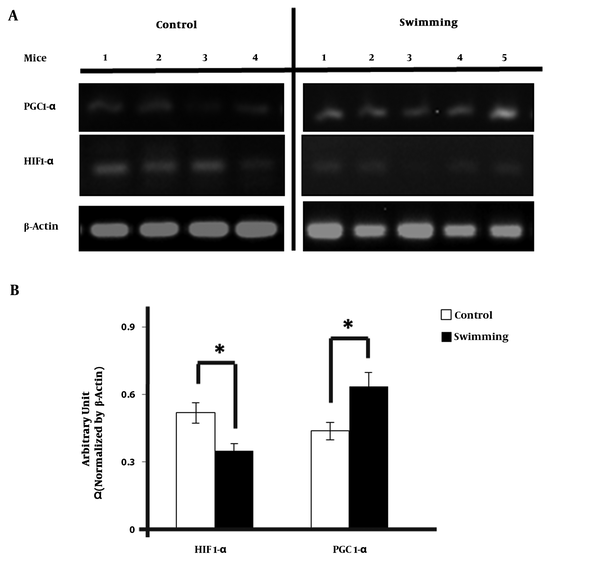
We examined the change in HIF1-α and PGC-1α expression in heart of mice by semi quantitative real time PCR method. PCR band of HIF1-α and PGC-1α (Figure 2A) were normalized using β-actin and quantified using ImageJ. HIF1-α expression in swimming group were significantly lower than the control group. The mean of the ratio was 0.64 in control group versus 0.44 in the exercise group (Figure 2B). The results were significantly higher compared with control group. (CI = 95%, -0.378 - 0.019; P = 0.035). But, after HIF-1α - β-actin gene ratio was obtained, it expression was significantly lower than control group with the mean of the ratio was 0.35 vs 0.52; (CI = 95%, 0.27 - 0.31; P = 0.025)
4. Discussion
Aerobic exercise includes jogging or running, Nordic walking, brisk walking, hiking, cycling, cross-country skiing, aerobic dancing, skating, rowing or swimming. It involves rhythmic movements of large muscle mass for a continued period (3). Swimming is a high stress training exercise which is frequently used as an exercise model for studying physiology mechanism in small laboratory animals. It has several advantages compared to other types of exercise such as running (2, 11). Animals in swimming protocols are often swimming spontaneously, without any aversive stimulation. So that, concentrations of stress hormones such as catecholamine is more stable compared with running protocols (11).
On the other hand, aerobic exercise by swimming has been shown to increase oxygen metabolism to produce ATP which is used in muscle contraction, therefore leading to enhanced formation of reactive oxygen species (ROS). It could reduce cardiac antioxidants to compensate this condition (12). An imbalance between production of ROS and antioxidant as counteraction can induce oxidative damage (13). It can damage the intact DNA strands, lipid oxidation and protein oxidation that could cause catastrophic cellular malfunctioning. Nevertheless due to the threat of oxidative damage, ROS is recognized as a signal for facilitating beneficial adaptations in affected tissue by cellular mechanism (14, 15). Thus, it seems possible that elevated ROS production in acute conditions, such as exercise, can trigger a protective status by signaling for increases in endogenous antioxidant enzymes through activation of redox-sensitive transcription factor (16, 17). In addition to increased ROS production, it stimulates PGC-1α expression as one of key the proteins in the pathway of mitochondrial biogenesis to provide energy that is needed in contraction (14, 18).
PGC-1α is an auxiliary transcription activating factor (transcriptional coactivator) that modulates of energy metabolism. It has also been proposed as a key mediator of long-term adaptation to exercise that powerfully regulates mitochondrial biogenesis, oxidative phosphorylation, and respiration (9, 19).
Our results are comparable to Terada et al. who performed a 6-hour acute bout of low intensity swimming exercise in 4 - 5-week-old Sprague-Dawley male rats. It showed that PGC-1α mRNA expression in epitrochlearis muscle increased approximately 8-fold compared to control group (20). Similarly in our research, four weeks of long-term of moderate intensity swimming exercise increased the gene expression of PGC-1α in cardiac muscle. Increased expression of these genes has been suggested to occur in response to altered energy demands that lead to hypoxia.
Other conditions showed that elevated PGC-1α regulates the stabilization of HIF-1 in skeletal muscle (21). HIF-1 is a transcriptional factor that modulates several genes of oxygen homeostasis in the state of oxygen deprivation, such as erythropoietin (EPO) and VEGF (22). Hypoxia condition leads to increase in the ionizing radiation, angiopoietin-2 and environmental stress that is induced by production of ROS. ROS may reduce Fe2+ availability, which inhibits the activity of prolyl hydroxylase domain-containing (PHD) protein and factor inhibiting HIF-1 (FIH-1). Both enzymes have function as regulators of HIF-1 and their activity requires oxygen as co-substrate (23). Inhibition of the PHD leads to HIF-1α stabilization to bind the coactivator CREB-binding protein (CBP)/p300 and continues the transcription activity (7, 24). Then, HIF-1α is accumulated in the cytoplasm under hypoxic condition rapidly and modified by sumoylation, s-nitrosylation, acetylation and phosphorylation (24). After phosphorylation, it subsequently translocates into the nucleus for dimerization with its partner HIF-1β forming the transcription factor HIF-1. Through the activation of HIF-1, it mediates the primary cellular responses to oxygen deprivation, promotes expression of several genes which encodes growth factor, transporters and enzymes that have a role in both short-and long-term adaptation to hypoxia (7, 24). HIF rapidly increases O2 supply through upregulation of nitric oxide (NO), the enzymatic product of inducible nitric oxide synthase (iNOS) that relaxes vascular smooth muscle cells, providing a short-term increase in blood flow. If short-term adaptation can not compensate the oxygen deprivation, it could stimulate angiogenesis as long-term adaptation (24). As stated in the previous study, HIF-1 alpha expression has also been associated with the presence of coronary collaterals in patients with coronary arterial disease (CAD). Therefore, both adaptations simultaneously increase the oxygen supply to the system and diminish the dependence of the cells on oxygen (25).
In research conducted by Flora et al. who used a rat treadmill with a specific exercise programs; HIF-1α proteins were increased in the groups with aerobic and anaerobic exercise. Concentrations of HIF-1α in myocardium were increased in the groups with aerobic and anaerobic exercise. The HIF 1α expression was highest in anaerobic exercise group which ran on treadmill for short term duration (7).
Contrarily, in our model of long-term moderate swimming exercise, the gene expression of HIF-1α in exercise group was lower than control group. This implies that decrease of HIF-1α expression is a major adaptation process that happens in the myocardium and is beneficial for oxidative capacity and endurance performance. Certain levels of HIF-1α are maintained in cardiac muscle not only under hypoxic but also in the normoxic condition of the control group if it is necessary to maintain metabolic functions. Without maintenance of HIF-1α, ATP production probably decreases and causes dysfunction of cardiac contractility.
Downregulation of HIF-1 activity with long-term aerobic exercise could be caused by negative regulators, such as FIH-1 and sirtuin (6). The FIH prevents binding to CBP/p300 through hydroxylation of an aspargine residue, while sirtuin 6 is a histone 3 lysine 9 deacetylase that epigenetically inhibits activation of the glycolytic response genes (26). Research conducted by Lindholm et al. showed that a longitudinal 6 week training programme increased expression of those negative regulators, indicating that the high levels of expression are a consequence of long-term exercise (27). Previous study also showed that chronic hypoxia of high altitude for generations, has a very high frequency of a variant of PHD2 with a higher affinity for oxygen (28).
As stated above, PGC-1α and HIF-1α are factors that contribute to conserving energy during the metabolic process. Alteration of both factors after moderate intensity of exercise could increase the capillarization, glycogen storage as well as altered glycolytic flux and mitochondrial density (26). These effects could improve metabolic capacity and oxygen supply to cardiac muscle and may be a beneficial mechanism to improve health in cardiovascular disease.
Future studies are needed to compare effects of different intensity exercises and the changes of cardiac function during and after training period. Future research, which provides a complete pathway of molecular mechanism that contributes to the metabolic adaptation process after aerobic exercise are needed.
4.1. Conclusions
Taken together, ST increased the expression of PGC-1α but decreased the expression of HIF-1α in mice cardiac muscle in response to chronic hypoxia condition. Alteration of these gene expressions may contribute to cardiac physiological adaptation during training.