1. Background
Heart rate variability (HRV) is a term used to describe the RR-interval oscillation, which is primarily due to both parasympathetic and sympathetic neural control of the heart. It is believed that HRV is a useful noninvasive indicator of cardiac autonomic function with important prognostic value (1).
In sports, an increase in HRV is associated with a high ability of the body to adapt to high physical stimuli (2). On the contrary, a decrease in HRV indices during growing physical exercises indicates incomplete recovery after intensive exercises and even overreaching state (3).
It has been shown that HRV of highly trained athletes is increased (4). Mechanisms of a HRV increase are not fully known. In addition to physical training effects, genetic predisposition to increased HRV is highly important (5). Twin studies show that the inheritance of HRV indexes is about 46% - 57% (5). However, the genetic polymorphisms causing increased HRV remain completely unclear.
Knowledge of molecular genetic factors affecting HRV indices will help to explain consistently high or low HRV during frequent HRV monitoring, as well as to improve the determination of fitness.
Genetic polymorphisms of the uncoupling proteins 2 (UCP2) and 3 (UCP3) are of particular interest regarding genetic factors determining autonomic regulation of the athletes’ hearts. These genes determine the synthesis and activity of uncoupling proteins of the appropriate type that can dissociate oxidation and phosphorylation in mitochondria of different tissues, that is, to change oxidations to increase heat generation, thereby reducing the metabolic efficiency of mitochondria (6, 7). In addition, UCP2 and UCP3 proteins are implicated in the formation of reactive oxygen species (ROS) by the mitochondria during respiration, and lowering of the proton gradient across the inner mitochondrial membrane by UCPs results in a reduction in ROS generation (7). Furthermore, these proteins affect insulin sensitivity (8), leptin level (9) and angiotensin-converting enzyme expression (10). All these functions of UCP2/3 may mediate the effects of these genetic polymorphisms on the autonomous regulation of the heart. Moreover, it was previously established that UCP2 45 bp I/D and UCP3-55C/T polymorphisms were associated with HRV and blood pressure in untrained men.
2. Objectives
The purpose of this study was to evaluate possible associations of the UCP2 Ala55Val and UCP3-55T/C polymorphisms with HRV of the trained oarsmen. This was a pilot study of the role of the UCP2/3 genes in the determination of HRV.
3. Methods
3.1. Subjects
Young healthy oarsmen (n = 23) who have been rowing regularly for more than 1.5 years in St. Petersburg sports clubs and participating in local and national competitions were invited to participate in the study. The age of the oarsmen was 17.6 ± 1.6 years old; their height: 180.2 ± 8.4 cm; body weight: 75.5 ± 10.4 kg, sports experience of rowing: 4.8 ± 2.9 years, VO2max: 55.4 ± 10.4 mL/min/kg. In the last month before the examination, the athletes trained according to a similar training program so that their relative intensity of loads was about the same, which caused similar changes in the autonomous regulation of the heart. Athletes did not have any abnormalities in the health and regulation of the cardiovascular system. The examination was carried out at the end of the pre-competition period with high training loads.
The study conforms to the code of ethics of the World Medical Association Declaration of Helsinki and ethical standards in sport and exercise science research. All underage (age < 18 years, n = 12) athletes’ parents and adult athletes gave their written consent after a detailed explanation about the study procedure and risks involved with the investigation.
3.2. Maximum Oxygen Consumption (VO2max) Test
VO2max was determined using an incremental test to exhaustion on a rowing ergometer Weba Sport Slider Kayak ergometer (Austria). The initial stage of the workload was 120 W and increased every two minutes by 75 W. The rest interval between the steps was 30 seconds. Oxygen consumption was recorded by means of a gas analyzer MetaMax 3B (Cortex, Leipzig, Germany). The heart rate was determined using a Polar RS 400sd heart rate monitor. Using the gas analyzer, O2 and CO2 contents were measured by the electrochemical and non-dispersive infrared sensors, respectively. The air flow was measured by a turbine transducer. Standard calibrations of gas sensors and transducer were performed before each test. Maximal oxygen consumption was recorded as the highest mean value recorded in the test observed over a 30s period in the final part of the workloads. The criteria used to confirm a maximal test were a decrease in power of more than 30 W from the target power; a respiratory exchange ratio greater than 1.1 and a final heart rate above 95% of the age-related maximum.
3.3. Genotyping
DNA analysis was performed in a specialized genetic laboratory. DNA was extracted from buccal cells using a DNK-sorb-A sorbent kit according to the manufacturer’s instruction (Central Research Institute of Epidemiology, Moscow, Russia).
UCP2 Ala55Val (rs660339) and UCP3-55T/C (rs1800849) polymorphisms were genotyped by the polymerase chain reaction-restriction fragment length polymorphism method in accordance with studies (11). The sequences of the primers (The Scientific Production Company “LITECH”. Moscow) to detect Ala55Val polymorphism were forward 5’-CTGGGAGTCTTGATGGTGTCTAC-3’ and reverse 5’-CACCGCGGTACTGGGCGTTG-3’, those to detect -55C/T polymorphism were forward 5’-GAGCTATATTAAAGCACCCCAAGT-3’ and reverse 5’-TCTGCTGCTTCTGGCTTGGCACTGGTCTTATACACCC-3’. The polymerase chain reactions were carried out as follows. Initial denaturation at 95°C for 5 minutes (1 cycle) followed by 30 cycles of denaturation at 95°C for 60 seconds, annealing at 63°C for 60 seconds, extension at 72°C for 60 seconds and a final extension at 72°C for 5 minutes. Amplicons (199 bp DNA fragments for UCP2; 189 bp DNA fragments for UCP3) were incubated (for UCP2 at 37°C and for UCP3 at 30°C during the night) together with restriction endonucleases (HindII for UCP2, SmaI for UCP3 produced by “SibEnzyme”. Novosibirsk. Russia) to identify single nucleotide replacements. Length analysis of restriction products was carried out by electrophoretic separation of 8% polyacrylamide gel followed by staining with ethidium bromide and visualization in ultraviolet light with a transilluminator. Unrestricted fragments of length 199 bp corresponded to the UCP2 Ala/Ala genotype, three fragments of length 199, 180, and 19 bp corresponded to the Ala/Val genotype and two fragments of length 180 and 19 bp - the Val/Val genotype. The UCP3 T/T genotype corresponded to an unrestricted fragment 189 bp long, the CT genotype three fragments 189, 152, and 37 bp long, and the CC genotype two fragments 152 and 37 bp long.
3.4. Analysis of HRV, Cardiac (CI) and Stroke (SI) Indices
Cardiohemodynamic measurements were performed in standard comfort conditions of a single room. After 5 minutes of supine rest, a lead II ECG and tetrapolar rheocardiogram (the active electrodes were placed on the jugular fossa and the xiphoid process) using the impedance cardiography hardware-software complex “Reodin-504” (Medass, Moscow) were simultaneously recorded during spontaneous breathing for 5 minutes in the supine position and during 6 minutes of active orthostasis. The first minute of orthostasis was excluded from the recordings to remove transient processes. Heart rate (HR), SI (mL/m2), CI (L/min/m2) were calculated using ECG and rheocardiograms.
To estimate HRV, we defined time-domain indices: standard deviations of NN intervals (SDNN), root mean square of successive differences (RMSSD), and power spectral measures (based on the fast Fourier transformation analysis): high-frequency (0.15 - 0.4 Hz, HF, ms2), low-frequency (0.04 - 0.14 Hz, LF, ms2) and very low-frequency (less than 0.04 Hz, VLF, ms2) powers of the RR-intervals oscillations. The normalized powers of HFnu, LFnu as well as LF/HF ratio were calculated (1). “Orto” was added to all studied indexes in order to mark the orthostatic position.
3.5. Statistics
The results are presented as the arithmetic mean ± standard deviation (SD). The correspondence of the genotypes distribution to the Hardy-Weinberg equilibrium was determined by comparing the observed and expected frequencies by the chi-square test. The associations of HRV, CI, and SI with the studied polymorphisms were assessed by a one-way ANOVA with the Bonferroni criterion for post-hoc comparisons. Student’s t-test was used for pair comparisons between the groups. Before the ANOVA analysis, HRV indices were converted by logarithm with a base of 10 (Lg).
4. Results
4.1. Distribution of the Athletes’ Genotypes and Alleles
Genotype distributions of UCP2 Ala55Val polymorphism (chi-square = 2.27, P = 0.321) and UCP3-55C/T polymorphism (chi-square = 0.631, P = 0.729) in all oarsmen (n = 23) were in Hardy-Weinberg equilibrium.
4.2. Associations of HRV and CI Indices with the UCP2 Ala55Val Polymorphism
4.2.1. The Supine Position
The UCP2 Ala55Val polymorphism was associated to HR (P = 0.001), SDNN (P = 0.006), RMSSD (P = 0.001), HF (P = 0.009), LF (P = 0.004), HFnu (P = 0.047), LF/HF (P = 0.044) and CI (P = 0.027), but an association with VO2max was not significant (P = 0.078) (Table 1). Major differences were connected with a lower HR and higher indices of HRV in persons with the Val/Val genotype regarding to the Ala/Val and (Ala/Ala + Ala/Val) groups.
| UCP2 Ala55Val Genotype | Post-Hoc | ||||||
|---|---|---|---|---|---|---|---|
| Ala/Ala (N = 3) | Ala/Val (N = 15) | Val/Val (N = 5) | P Value 1c | P Value 2d | P Value 3e | P Value 4f | |
| VO2max, mL/min/kg | 50.5 ± 8.4 | 53.4 ± 9.0 | 64.3 ± 11.7 | 0.078 | 1.00 | 0.185 | 0.116 |
| Lg HR, Lg bpm | 1.79 ± 0.07 | 1.84 ± 0.04 | 1.70 ± 0.07 | 0.001 | 0.448 | 0.058 | 0.001 |
| 1.83 ± 0.05 | 1.70 ± 0.07 | 0.001 | |||||
| Lg SDNN, Lg ms | 1.89 ± 0.04 | 1.71 ± 0.17 | 2.00 ± 0.16 | 0.006 | 0.266 | 1.000 | 0.007 |
| 1.74 ± 0.17 | 2.00 ± 0.16 | 0.006 | |||||
| Lg RMSSD, Lg ms | 1.78 ± 0.12 | 1.62 ± 0.15 | 2.01 ± 0.24 | 0.001 | 0.459 | 0.239 | 0.001 |
| 1.65 ± 0.16 | 2.01 ± 0.24 | 0.001 | |||||
| Lg HF, Lg ms2 | 3.16 ± 0.22 | 3.03 ± 0.43 | 3.74 ± 0.33 | 0.009 | 1.000 | 0.181 | 0.008 |
| 3.05 ± 0.40 | 3.74 ± 0.33 | 0.002 | |||||
| Lg LF, Lg ms2 | 3.52 ± 0.23 | 2.96 ± 0.34 | 3.49 ± 0.30 | 0.004 | 0.035 | 1.000 | 0.013 |
| 3.05 ± 0.38 | 3.49 ± 0.30 | 0.028 | |||||
| Lg VLF, Lg ms2 | 2.86 ± 0.42 | 2.84 ± 0.48 | 3.32 ± 0.43 | 0.149 | 1.000 | 0.566 | 0.171 |
| 2.84 ± 0.46 | 3.32 ± 0.43 | 0.048 | |||||
| Lg LF/HF, Lg % | 0.37 ± 0.32 | -0.07 ± 0.32 | -0.24 ± 0.26 | 0.044 | 0.114 | 0.043 | 0.890 |
| 0.00 ± 0.36 | -0.24 ± 0.26 | 0.168 | |||||
| HFnu, % | 31.5 ± 14.9 | 53.9 ± 17.0 | 62.8 ± 13.5 | 0.047 | 0.122 | 0.046 | 0.899 |
| 50.2 ± 18.4 | 62.8 ± 13.5 | 0.170 | |||||
| LFnu, % | 68.5 ± 14.9 | 46.1 ± 17.0 | 37.2 ± 13.5 | 0.047 | 0.122 | 0.046 | 0.899 |
| 49.8 ± 18.4 | 37.2 ± 13.5 | 0.170 | |||||
| CI, L/min/m2 | 3.52 ± 0.57 | 3.62 ± 0.75 | 2.60 ± 0.45 | 0.027 | 1.00 | 0.235 | 0.025 |
| 3.60 ± 0.70 | 2.60 ± 0.45 | 0.007 | |||||
| SI, mL/m2 | 57 ± 12 | 53 ± 11 | 53 ± 13 | 0.819 | 1.00 | 1.00 | 1.00 |
| 54 ± 11 | 53 ± 13 | 0.868 | |||||
Abbreviations: HR, heart rate; RMSSD, root mean square of successive differences; SDNN, standard deviations of NN intervals.
aValues are expressed as mean ± SD.
bLg, logarithm with a base of 10.
cSignificance level for ANOVA.
dAla/Ala vs. Ala/Val.
eAla/Ala vs. Val/Val.
fAla/Val vs Val/Val.
4.2.2. The Orthostatic Position
In the orthostatic position the UCP2 Ala55Val polymorphism was associated with HR (P = 0.022), SDNN (P = 0.006), RMSSD (P = 0.001) and CI (P = 0.029) (Table 2). The athletes with the UCP2 Val/Val genotype had decreased HR (P = 0.022), increased SDNN (P = 0.060) and RMSSD (P = 0.020), as well as low CI (P = 0.007) compared to the (AlaAla + Ala/Val) group. The UCP2 Ala55Val polymorphism was not associated with the responses of HRV, CI, and SI in response to orthostasis (all P > 0.1 results not shown).
| UCP2 Ala55Val Genotype | Post-Hoc | ||||||
|---|---|---|---|---|---|---|---|
| Ala/Ala (N = 3) | Ala/Val (N = 15) | Val/Val (N = 5) | P Value 1c | P Value 2d | P Value 3e | P Value 4f | |
| Lg HRortho, Lg bpm | 1.84 ± 0.08 | 1.90 ± 0.07 | 1.81±0.06 | 0.022 | 0.412 | 1.00 | 0.027 |
| 1.89 ± 0.07 | 1.81 ± 0.06 | 0.019 | |||||
| Lg SDNNortho, Lg ms | 1.83 ± 0.13 | 1.63 ± 0.13 | 1.83 ± 0.13 | 0.006 | 0.056 | 1.00 | 0.018 |
| 1.66 ± 0.15 | 1.83 ± 0.13 | 0.030 | |||||
| Lg HFortho, Lg ms2 | 2.83 ± 0.52 | 2.73 ± 0.49 | 3.03 ± 0.30 | 0.474 | 1.00 | 1.00 | 0.685 |
| 2.75 ± 0.48 | 3.03 ± 0.30 | 0.235 | |||||
| Lg LFortho, Lg ms2 | 3.46 ± 0.39 | 3.14 ± 0.41 | 3.33 ± 0.37 | 0.379 | 0.668 | 1.00 | 1.00 |
| 3.19 ± 0.41 | 3.33 ± 0.37 | 0.513 | |||||
| Lg VLFortho, Lg ms2 | 2.72 ± 0.15 | 2.68 ± 0.38 | 2.97 ± 0.18 | 0.241 | 1.00 | 0.885 | 0.291 |
| 2.69 ± 0.34 | 2.97 ± 0.18 | 0.089 | |||||
| Lg LF/Hfortho, Lg % | 0.63 ± 0.59 | 0.41 ± 0.37 | 0.30 ± 0.47 | 0.570 | 1.00 | 0.888 | 1.00 |
| 0.44 ± 0.40 | 0.30 ± 0.47 | 0.500 | |||||
| Lg RMSSDortho, Lg ms | 1.65 ± 0.21 | 1.38 ± 0.15 | 1.70 ± 0.17 | 0.001 | 0.041 | 1.00 | 0.003 |
| 1.42 ± 0.19 | 1.70 ± 0.17 | 0.008 | |||||
| HFnu(ortho) | 24.3 ± 21.1 | 30.5 ± 16.4 | 36.3 ± 22.2 | 0.665 | 1.00 | 1.00 | 1.00 |
| 29.5 ± 16.7 | 36.3 ± 22.2 | 0.462 | |||||
| LFnu(ortho) | 75.7 ± 21.1 | 69.5 ± 16.4 | 63.7 ± 22.2 | 0.665 | 1.00 | 1.00 | 1.00 |
| 70.5 ± 16.7 | 63.7 ± 22.2 | 0.462 | |||||
| CIortho, L/min/m2 | 2.94 ± 0.78 | 2.86 ± 0.82 | 1.75 ± 0.54 | 0.029 | 1.00 | 0.139 | 0.032 |
| 2.87 ± 0.79 | 1.75 ± 0.54 | 0.008 | |||||
| SIortho, mL/m2 | 43 ± 15 | 36 ± 10 | 28 ± 11 | 0.156 | 0.814 | 0.187 | 0.523 |
| 37 ± 11 | 28 ± 11 | 0.111 | |||||
Abbreviations: HR, heart rate; RMSSD, root mean square of successive differences; SDNN, standard deviations of NN intervals.
aValues are expressed as mean ± SD.
bLg, logarithm with a base of 10.
cSignificance level for ANOVA.
dAla/Ala vs. Ala/Val.
eAla/Ala vs. Val/Val.
fAla/Val vs Val/Val.
4.3. Associations of HRV and CI Indices with the UCP3-55T/C Polymorphism
4.3.1. The Supine Position
The UCP3-55T/C polymorphism was associated with HR (P = 0.001), RMSSD (P = 0.006), HF (P = 0.032), LF (P = 0.053), VLF (P = 0.047) and SI (P = 0.085), there was not a correlation with VO2max (Table 3). The T/T genotype had lower HR (P = 0.001), and higher levels of SDNN (P = 0.007), RMSSD (P = 0.002), HF (P = 0.008), LF (P = 0.014), VLF (P = 0.013) compared to the (C/T + C/C) group. Comparison of the C/C genotype with the (C/T + T/T) group did not reveal any significant differences (all P > 0.1 results not shown).
| UCP3-55T/C Genotype | Post-Hoc | ||||||
|---|---|---|---|---|---|---|---|
| TT (N = 3) | CT(N = 13) | CC (N = 7) | P Value 1c | P Value 2d | P Value 3e | P Value 4f | |
| VO2max, mL/min/kg | 63.5 ± 1.2 | 56.0 ± 12.3 | 50.7 ± 5.5 | 0.196 | 0.758 | 0.237 | 0.833 |
| Lg HR, Lg bpm | 1.66 ± 0.06 | 1.82 ± 0.05 | 1.81 ± 0.05 | 0.001 | 0.001 | 0.001 | 1.00 |
| 1.66 ± 0.06 | 1.82 ± 0.05 | 0.001 | |||||
| Lg RMSSD, Lg ms | 2.08 ± 0.14 | 1.65 ± 0.014 | 1.73 ± 0.27 | 0.006 | 0.005 | 0.039 | 1.00 |
| 2.08 ± 0.14 | 1.68 ± 0.19 | 0.002 | |||||
| Lg SDNN, Lg ms | 2.07 ± 0.13 | 1.75 ± 0.017 | 1.78 ± 0.19 | 0.027 | 0.025 | 0.070 | 1.00 |
| 2.07 ± 0.13 | 1.76 ± 0.17 | 0.007 | |||||
| Lg HF, Lg ms2 | 3.85 ± 0.019 | 3.11 ± 0.43 | 3.08 ± 0.47 | 0.032 | 0.039 | 0.048 | 1.00 |
| 3.85 ± 0.019 | 3.10 ± 0.43 | 0.008 | |||||
| Lg LF, Lg ms2 | 3.67 ± 0.06 | 3.06 ± 0.32 | 3.08 ± 0.049 | 0.053 | 0.056 | 0.101 | 1.00 |
| 3.67 ± 0.06 | 3.07 ± 0.38 | 0.014 | |||||
| Lg VLF, Lg ms2 | 3.57 ± 0.51 | 2.88 ± 0.46 | 2.80 ± 0.35 | 0.047 | 0.069 | 0.058 | 1.00 |
| 3.57 ± 0.51 | 2.85 ± 0.42 | 0.013 | |||||
| Lg LF/HF, Lg % | -0.18 ± 0.15 | -0.05 ± 0.28 | 0.01 ± 0.051 | 0.749 | 1.00 | 1.00 | 1.00 |
| -0.18 ± 0.15 | -0.03 ± 0.37 | 0.488 | |||||
| HFnu, % | 60.3 ± 8.3 | 53.0 ± 15.3 | 49.7 ± 25.6 | 0.713 | 1.00 | 1.00 | 1.00 |
| 60.3 ± 8.3 | 51.8 ± 18.9 | 0.460 | |||||
| LFnu, % | 39.7 ± 8.3 | 47.0 ± 15.3 | 50.3 ± 25.6 | 0.713 | 1.00 | 1.00 | 1.00 |
| 39.7 ± 8.3 | 48.2 ± 18.9 | 0.460 | |||||
| CI, L/min/m2 | 2.86 ± 0.40 | 3.32 ± 0.72 | 3.72 ± 0.91 | 0.262 | 1.00 | 0.354 | 0.828 |
| 2.86 ± 0.40 | 3.46 ± 0.79 | 0.221 | |||||
| SI, mL/m2 | 63 ± 10 | 49 ± 9 | 57 ± 13 | 0.085 | 0.161 | 1.00 | 0.338 |
| 63 ± 10 | 52 ± 11 | 0.121 | |||||
Abbreviations: HR, heart rate; RMSSD, root mean square of successive differences; SDNN, standard deviations of NN intervals.
aValues are expressed as mean ± SD.
bLg, logarithm with a base of 10.
cSignificance level of differences for ANOVA.
dT/T vs. C/T.
eT/T vs. C/C.
fC/T vs. C/C.
4.3.2. The Orthostatic Position
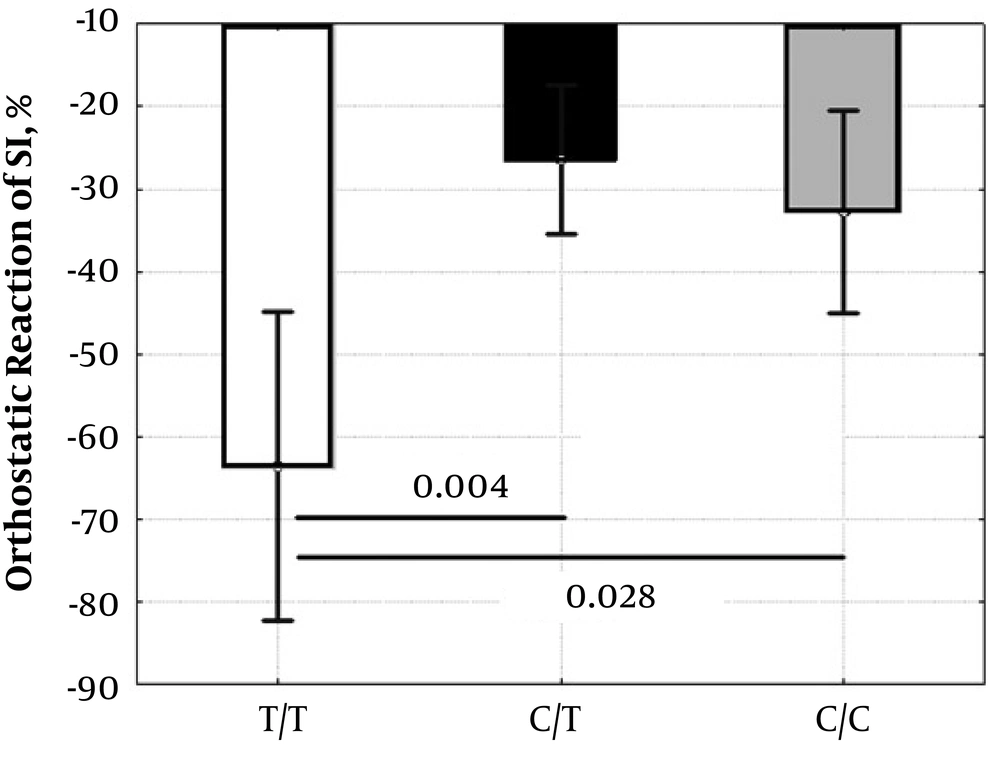
In the standing position, the UCP3-55T/C polymorphism was associated only with LF (P = 0.024), and the indices CIorto (P = 0.055), SIorto (P = 0.089), SDNNorto (P = 0.063) had near significant associations (Table 4). However, the UCP3-55T/C polymorphism showed associations with HR response (mean ± SD: T/T = 59 ± 11%, C/T = 10 ± 15%, C/C = 24 ± 7%, ANOVA: P < 0.001; T/T vs. C/T P < 0.001; T/T vs. C/C P = 0.002; C/T vs. C/C P = 0.122) and SI response (ANOVA: P = 0.005; Figure 1 ) to active orthostasis.
| UCP3-55T/C Genotype | Post-Hoc | ||||||
|---|---|---|---|---|---|---|---|
| TT (N = 3) | CT (N = 13) | CC (N = 7) | P Value 1c | P Value 2d | P Value 3e | P Value 4f | |
| Lg HRortho, Lg bpm | 1.86 ± 0.04 | 1.86 ± 0.09 | 1.90 ± 0.06 | 0.546 | 1.00 | 1.00 | 0.902 |
| 1.86 ± 0.04 | 1.88 ± 0.08 | 0.737 | |||||
| Lg RMSSDortho, Lg ms | 1.56 ± 0.28 | 1.43 ± 0.18 | 1.55 ± 0.24 | 0.408 | 1.00 | 1.00 | 0.741 |
| 1.56 ± 0.28 | 1.47 ± 0.21 | 0.512 | |||||
| Lg SDNNortho, Lg ms | 1.83 ± 0.16 | 1.63 ± 0.14 | 1.76 ± 0.15 | 0.063 | 0.134 | 1.00 | 0.245 |
| 1.83 ± 0.16 | 1.68 ± 0.15 | 0.116 | |||||
| Lg HFortho, Lg ms2 | 3.12 ± 0.33 | 2.78 ± 0.47 | 2.74 ± 0.48 | 0.474 | 0.806 | 0.744 | 1.00 |
| 3.12 ± 0.33 | 2.76 ± 0.46 | 0.222 | |||||
| Lg LFortho, Lg ms2 | 3.75 ± 0.09 | 3.08 ± 0.36 | 3.26 ± 0.38 | 0.024 | 0.022 | 0.166 | 0.858 |
| 3.75 ± 0.09 | 3.14 ± 0.37 | 0.011 | |||||
| Lg VLFortho, Lg ms2 | 3.00 ± 0.05 | 2.71 ± 0.40 | 2.71 ± 0.24 | 0.391 | 0.579 | 0.654 | 1.00 |
| 3.00 ± 0.05 | 2.71 ± 0.34 | 0.166 | |||||
| Lg LF/Hfortho, Lg % | 0.63 ± 0.24 | 0.30 ± 0.37 | 0.52 ± 0.50 | 0.318 | 0.636 | 1.00 | 0.760 |
| 0.63 ± 0.24 | 0.38 ± 0.42 | 0.323 | |||||
| Lg RMSSDortho, Lg ms | 1.56 ± 0.28 | 1.43 ± 0.18 | 1.55 ± 0.24 | 0.408 | 1.00 | 1.00 | 0.741 |
| 1.56 ± 0.28 | 1.47 ± 0.21 | 0.512 | |||||
| HFnu(ortho) | 19.8 ± 9.3 | 35.3 ± 17.7 | 27.7 ± 19.5 | 0.351 | 0.558 | 1.00 | 1.00 |
| 19.8 ± 9.3 | 32.6 ± 18.2 | 0.251 | |||||
| LFnu(ortho) | 80.2 ± 9.3 | 64.7 ± 17.7 | 72.3 ± 19.5 | 0.351 | 0.558 | 1.00 | 1.00 |
| 80.2 ± 9.3 | 67.4 ± 18.2 | 0.251 | |||||
| CIortho, L/min/m2 | 1.68 ± 0.90 | 2.60 ± 0.65 | 3.09 ± 0.97 | 0.055 | 0.257 | 0.053 | 0.593 |
| 1.68 ± 0.90 | 2.77 ± 0.79 | 0.041 | |||||
| SIortho, mL/m2 | 23 ± 10 | 36 ± 9 | 39 ± 13 | 0.089 | 0.184 | 0.095 | 1.00 |
| 23 ± 10 | 37 ± 10 | 0.034 | |||||
Abbreviations: HR, heart rate; RMSSD, root mean square of successive differences; SDNN, standard deviations of NN intervals.
aValues are expressed as mean ± SD.
bLg, logarithm with a base of 10.
cSignificance level of differences for ANOVA.
dT/T vs. C/T.
eT/T vs. C/C.
fC/T vs. C/C.
5. Discussion
This study evaluated the associations of the UCP2 Ala55Val and the UCP3-55C/T polymorphisms with HRV and cardiac index (CI) in supine and standing positions in the young oarsmen. Our main findings showed that higher HRV and lower HR was associated with UCP2 Val/Val and UCP3 T/T genotypes in a supine position and less significant in orthostasis.
It is believed that the UCP2 Val/Val genotype is associated with low uncoupling activity of mitochondria, increased metabolic efficiency during rest and exercise, reduced fat oxidation and higher spontaneous physical activity (12, 13). Although we did not reveal a significant correlation of the polymorphism with aerobic capacity, however, there was a tendency for increased VO2max (P = 0.078) in the Val/Val genotype, which is consistent with the works (11, 13, 14), which showed the positive effect of the Val allele on physical performance.
The results of our study showed that the positive effect of the Val/Val genotype may be associated with the activation of cardiac vagus, because athletes with Val/Val genotype had higher HRV.
The mechanism of the associations is poorly understood, since reduced UCP2 activity associated with Val/Val genotype is more likely correlated with increased ROS formation (7), and higher activity of the angiotensin-converting enzyme (10) which have sympathetic effects. It is known that higher ROS levels (15) and activity of the renin-angiotensin system (16) are associated with an increased sympathetic activity and decreased HRV. It is also shown (17) that UCP2 45 bp I/I genotype having low UCP2 activity was associated with lower HRV.
We suppose that in our study increased cardiac parasympathetic activities in the Val/Val genotype group may be due to higher insulin sensitivity (8) or lower leptin levels (9). In some studies were shown that both factors are associated to Val/Val genotype and increased HRV indices (18, 19). However, one cannot exclude the possibility that the associations of Val allele to higher HRV could be due to other polymorphisms that is in the linkage disequilibrium with Ala55Val polymorphism (20). Generally, physiologic mechanisms of increased parasympathetic activities in the Val/Val genotype carriers require further confirmation and research.
Besides the UCP2 Val/Val genotype, the UCP3-55T/T genotype was associated to lower HR and increased HRV in our athletes. In contrast to the UCP2 Val/Val genotype, the UCP3-55T/T genotype contributes to the higher expression of UCP3 generally in muscles disrupting oxidation and phosphorylation inducing decreased ATP synthesis and ROS formation (21).
A number of mechanisms can be proposed to explain increased HRV in athletes with the UCP3 T/T genotype. Increased cardiac vagus tone in UCP3 T/T athletes may be associated with low angiotensin 2 activity, increased insulin sensitivity or low ROS levels. Firstly, it was shown that angiotensin-converting enzyme activity (10), blood glucose and insulin resistance (8) and presumably tissue ROS levels (7) may be reduced in carriers of the UCP3 T/T genotype. Secondly, reduced angiotensin 2 levels (16), high insulin sensitivity (19) and low ROS levels (15), as a rule, are correlated with elevated HRV. Moreover, similar associations between HRV and UCP3-55T/C polymorphism have been established in the work (17) which showed increased parasympathetic HRV indices in carriers of the -55T allele compared to carriers of the UCP3 C/C genotype. However, all these mechanisms are speculative and require further research.
Another feature of the UCP3-55T/C polymorphism is the association of the T/T genotype with an increased response of HR and SI to orthostasis. This may indicate orthostatic intolerance in athletes with the T/T genotype. We cannot specify the effects of the UCP3 T/T genotype, which may contribute to a decrease in orthostatic stability. However, taking into account the mechanisms of development of orthostatic intolerance among athletes (22), it can be assumed that the involvement of the UCP3 T/T genotype in reducing ROS and angiotensin 2 levels, linked anti-inflammatory activity and other pleiotropic effects of this genotype may contribute to increased dilatation and compliance of the vascular system and myocardium (eccentric hypertrophy) (23) and lead to a side orthostatic intolerance in athletes with the T/T genotype. Thus, we believe that the associations of the T/T genotype with higher SI and HR orthostatic responses may reflect the involvement of this genotype in increasing compliance and dilatation of the vascular wall and myocardium induced by physical exercises.
Despite the lack of associations between the UCP2 and UCP3 polymorphisms with VO2max, our data are consistent with the works (11, 14) and indicate that UCP2 55Val and UCP3-55T alleles can positively affect athletes’ adaptation to high physical exertion, because these alleles were associated with higher cardiac vagus tone, which is a favorable marker of high athletic performance.
5.1. Limitations
Due to the small number of the athletes surveyed, especially with UCP2 Ala/Ala and UCP3 T/T genotypes, the statistical power of this study is moderate, so, at the current time it requires caution when generalizing the results obtained. In addition, further studies with a larger number of subjects are required to understand the associations between UCP2 and UCP3 polymorphisms and cardiac autonomic regulation.
5.2. Conclusions
The findings of our pilot study show that the athletes with the UCP2 Val/Val and the UCP3 T/T genotypes have increased HRV suggesting an influence of UCP2/UCP3 polymorphisms on autonomic cardiovascular regulation. Consequently, these genotypes, often found in athletes and associated with high athletic and physical performance, can at least in part constitute a genetic basis that explains the increased HRV in highly trained athletes. However, the role of the UCP2 Ala55Val and the UCP3-55T/C polymorphisms in determining HRV needs further clarification.