1. Background
Amongst the existing X-linked bleeding disorders, hemophilia A, with its recessive nature, is known as one of the most common ones. With the scientifically agreed-upon frequency of one in 5000 male births, the cause of this disorder is proved to be a sort of defect of the plasma protein involved in hemostasis called FVIII (1). Based on the natural function of FVIII, hemophilia A is subcategorized into three various levels of mild (> 5% - < 40%), moderate (1% - 5%) and severe (< 1%) (2), with the prevalence of 40%, 10%, and 50%, respectively (3). As an essentially functional plasma protein for the coagulation system of the human blood, FVIII is transformed into an inactive form via establishing a connection with the von Willebrand factor during the clotting process. When the body receives an injury, coagulation factor VIII activates and separates from the von Willebrand factor. The next step of the procedure of responding to injury is the interaction of factor VIII with another coagulation factor, factor IX. When these two factors interact with one another, a sequence of resultant chemical reactions leads to the coagulation of the blood (4).
With its exact location in the proximal segment of chromosome Xq28, the human FVIII (F8) gene has the function of encoding the coagulation factor VIII. Approximately, the length of this gene is 186 kb, including 26 exons, which leads to the production of two transcripts, each of which is spliced alternatively. A respectively large glycoprotein, known as isoform a, is encoded by the transcript variant 1; correspondingly, a putative small protein, isoform b, is encoded by the transcript variant 2 (5). More than thousand different mutations are reported in the F8 gene, including over 120 main deletions (< 50 bp); inversion in intron 1, which is accountable for 1% - 4% of patients with severe forms of hemophilia A; inversion in intron 22, which is accountable for nearly half of patients with severe forms of hemophilia A; and small substitutes and deletions of unspecified nature (6). More than 1000 mutations are archived thus far in the Hemophilia World Databank, also known as HAMSTeRs (7).
The microRNAs (miRNAs) are short non-coding RNAs with 18 - 25 nucleotides in length. Their predominant function is to work as the regulator of post-transcriptional gene expression.
Firstly, miRNAs are transcribed into the form of elongated primary transcripts, namely pri-miRNA. Next, being processed by the Drosha enzyme and then the precursor miRNA (pre-miRNA), it is moved to the cytoplasm. In the cytoplasm, it undergoes further processing by the Dicer enzyme. Eventually, the mature miRNA is integrated into an RNA-induced silencing complex (RISC), attached to the 3’ untranslated region (UTR) of the target messenger RNAs (mRNAs), specifically to mediate translational repression (8).
Allegedly, about 60% of all human genes are believed to be the reputed targets of single miRNAs since an individual miRNA is potent enough for targeting up to hundreds of genes. A complex regulatory network is additionally created since human genes might contain multiple binding sites for various miRNAs (9).
It is comprehensively elucidated that miRNAs are involved in various biological processes since they play a pivotal role in the pathogenesis process of different diseases and cancers.
The miRNA expression profiling signifies their tissue-specific patterns of expression; thus, they are considered as attractive biomarkers in diagnosis and treatment (10). Consequently, predicting and identifying parts of the genome that are susceptible to the expression of novel miRNAs, as well as registered miRNAs, leading to an outstanding opportunity for molecular studies of miRNAs, a target that can be acquired by both bioinformatics and molecular laboratory techniques (11).
2. Objectives
Hemophilia A is a monogenic disorder, and the majority of known and registered miRNAs in human genes influence the expression levels of its associated target gene; hence, the present study aimed at searching for the miRNAs embedded within the sequence of the F8 gene to control the progression of hemophilia A.
3. Methods
Based upon the sequences of known registered miRNAs, the types of algorithms that can be utilized in databases and bioinformatics tools to identify and predict novel miRNAs are initially pinpointed. Such programs are subsequently used to scan the genome and detect the sequences of putative novel miRNAs. In other words, gathering, analyzing, and combining a vast amount of data on known and registered miRNAs reveals similar characteristics, including the bulge size and position, the content of the nucleotide, the thermodynamic stability, sequence complexity, the length of the stem-loop structure and repetitive elements existing in the genes, which not only encode miRNAs but also are utilized in their prediction.
The current study employed the SSCprofiler database, and investigated two novel stem-loop structures in the F8 gene (http://mirna.imbb.forth.gr/SSCprofiler.html) (12).
Biological information concerning factors, such as structure, sequence, and protection of human miRNAs, is provided by the SSCprofiler database. On this database, the sensitivity and specificity of the output data are 84.16% and 95.88%, respectively (13). Moreover, the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (14) was utilized to study the stem-loop structures and stability produced by the SSCprofiler database. Processes, such as predicting secondary structures of single-stranded RNAs and calculating the base-pairing probability matrix, the partition function, and the structure of minimum free energy (MFE), are carried out by the RNAfold web server (15).
The following three web servers were used to assess the accuracy of the predicted stem-loop structures: miREval (http://mimirna.centenary.org.au/mireval) (16), FOMmiR (http://app.shenwei.me/cgi-bin/FOMmiR.cgi) (17), and MaturBayes (http://mirna.imbb.forth.gr/MatureBayes.html) (18). A support vector machine (SVM) trained on 57 distinct characteristics, such as sequence composition, secondary structure, and free energy, is utilized by the miREval web server. For the miREval database to differentiate miRNA stem-loops from stem-loops in other non-coding RNAs, two negative and positive categories of information are allocated to the SVM (16).
The FOMmiR database is capable of distinguishing the stem-loops from the precursors of miRNA, and also locates the position and strand of the mature miRNA. Therefore, this database represents a particular awareness upon biological recognition, which might be intently connected to the enzyme cleavage mechanism during the miRNA maturation (17). Besides, the MaturBayes database is a tool to detect mature miRNAs within the stem-loop structures, using a naive Bayes classifier.
Subsequently, the roles of the Dicer and Drosha enzymes were studied according to the stem-loop structure sequences, using the miR-FIND database (http://140.120.14.132:8080/MicroRNAProject-Web) (19).
Using the UCSC genome browser database (https://genome.ucsc.edu) (20), the conservation of the potential stem-loop structures in the vertebral genome was additionally analyzed. In 14 various human cell lines, the RNA expression profiling was investigated using deep sequencing technology; and the possibility of the novel miRNA expression in the potential sequence was assessed. Eventually, the miRBase database (http://www.mirbase.org/) (21) was utilized to authenticate the novelty of the potent sequences as mature miRNAs. Significantly, the miRNA gene potential candidates did not demonstrate an obvious sequence similarity to the known miRNA genes (22). On this database, over 12,000 mature miRNAs from 600 distinct species are identified and registered.
4. Results
To identify and predict the stem-loop structures, the F8 gene associated with hemophilia A was accordingly scanned. For this purpose, high reliable bioinformatics servers and related databases were utilized. Two stem-loop structures, namely, put-miR1 and put-miR2, with the sequences of “TGCTGCTGCCACTCAGGAAGAGGGTTGGAGTAGGCTAGGAA-TAGGAGCACAAATTAAAGCTCCTGTTCACTTTGACTTCTCC-ATCCCTCTCCTCCTTTCCTTAA”, and “TGTAAAAGGCTCATAAAAGTTGAGGAAGCCATTTGGGCTCtgctactccagcatggtccacagaccaggagtagcagcatcacctgagggcaattcaaaatgca”, respectively, both located in the first intron of the F8 gene, were predicted and presented for experimental verification, with regard to the results.
4.1. SSCprofiler Database
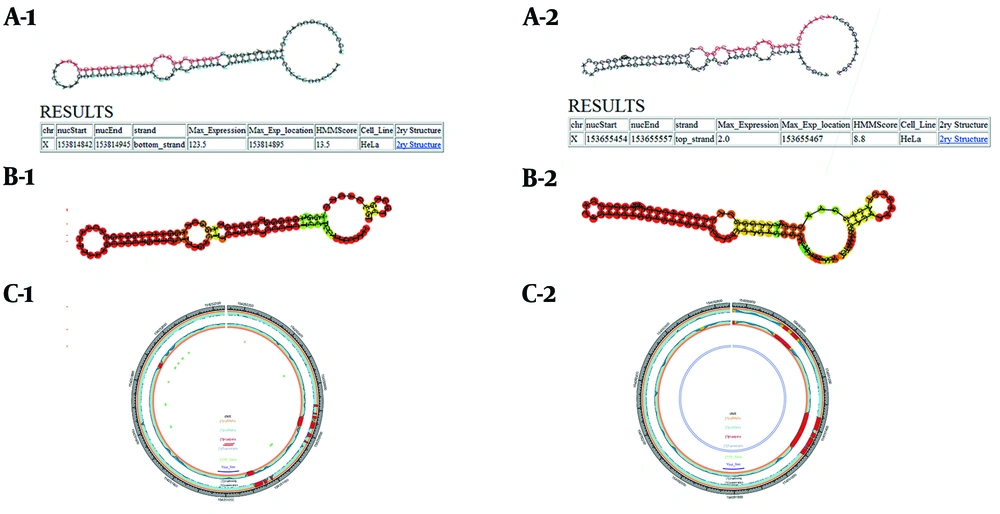
Stem-loop structures in the F8 gene are predicted and identified in this database. Moreover, the SSCprofiler database utilizes a hidden Markov model (HMM) to model secondary structural features in each position of miRNA stem-loops. The structure, sequencing, and conservation of the miRNA coding genes are simultaneously determined by this score in the statistical models; therefore, the higher the score, the greater the chance that the potential candidate structure belongs to the real miRNA (Figure 1A).
Using SSCprofiler, RNAfold, and miREval to predict put-miR1 and put-miR2 within the first intron of the human F8 gene, respectively. A-1, A-2: Results of SSCprofiler for put-miR1and put-miR2. Hairpin structures containing a probable sequence of mature miR (red) are shown, and HMM scores related to these structures are shown in the table. Furthermore, maximum expression (max-expression), according to a full genome tiling array in the HeLa cell line is presented for these sequences. B-1, B-2: Graphical output of hairpin structures in RNAfold web server. Secondary structure results of put-miR1 and put-miR2 are depicted. C-1, C-2: miREval output data; 1000 base pairs around our inquiry sequences are displayed as a circle graph by miREval.
4.2. RNAfold Web Server
To make more precise studies on the stability of the secondary structures, the stem-loop structures proposed by the SSCprofiler database were introduced in this server. Concerning the amount of minimum free energy apportioned amongst structures, the stability of the proposed secondary structures was investigated (Figure 1B). The MFE for put-miR1 and put-miR2 was -41.8 and -35.1 kcal.mol-1, respectively.
4.3. The miREval Web Tool
Through this web tool, the accuracy of the stem-loop structures was assessed (Figure 1C).
4.4. FOMmiR and MatureBayes Web Tools
Via FOMmiR and MatureBayes web tools, the predicted results for mature miRNAs in the candidate sequences, as well as the accuracy of the formation of secondary structures, were investigated (Figure 2A and B).
The results of FOMmiR, MatureBayes, and UCSC genome browser used to confirm the presence of novel microRNAs. A-1, A-2: the FOMmiR database information; the predicted mature miRNA sequence is observed in red in the candidate stem-loop structure. B-1, B-2: MatureBayes database output; the 3p and 5p sequences of mature miRNA are identified by a nucleotide position in the candidate sequence. C-1, C-2: Results of UCSC genome browser on human Feb.2009 (GRCH37/hg19) assembly. Conservation levels are shown with blue columns.
4.5. The miRFIND Database
Drosha and Dicer cleavage sites were identified in the candidate sequences and shown in Table 1.
| Sequence | Put-miR1 | Put-miR2 | ||
|---|---|---|---|---|
| Mature-miRNA Drosha/Dicer processing site | 23/46 | 82/59 | 20/42 | 79/58 |
| Mature-miRNA sequence | 5-UUGGAGUAGGCUAGGAAUAGGA-3 | 5-UCCUGUUCACUUUGACUUCUCCAU-3 | 1 5- AUGGAGAAGUCAAAGUGAACAGG-3 | 5-UCCUAUUCCUAGCCUACUCCAA-3 |
| Predicted seed site | 5-UGGAGUA-3 | 5-CCUGUUC-3 | 5-UGGAGAA-3 | 5-CCUAUUC-3 |
aData corresponding to the mature -5p (red) and miRNA-3p (black) are presented.
4.6. The UCSC Genome Browser
The percentage of sequences conservation among 100 vertebral genomes (Figure 2C), as well as the deep sequencing data (Figure 3), was analyzed. Conclusively, the results revealed that put-miR1 and put- miR2 were expressed in IMR90 CIP-TAP, IMR90, SKMC cells, and A549 CIP-TAP, HPC-PL TAP-only, IMR90 cells, respectively.
Deep sequencing information. The expression of the candidate sequences is represented by short RNAs (including miRNAs, etc.) in different cell types IMR90 CIP-TAP, IMR90, SKMC cells, and A549 CIP-TAP, HPC-PL TAP-only, IMR90 cells, in the put-miR1 and put- miR2, respectively. Regarding the expression pattern, it can be noted that the probability of mature miRNA presence in the candidate sequences associated with the F8 gene would increase.
4.7. The MiRBase Database
This database was utilized to ensure that the candidate sequences were not reported as a mature miRNA in other previously published studies.
5. Discussion
Nowadays, treatment decisions, as well as detection of recurrent disease and monitoring therapy, are mostly performed by predictive and diagnostic biomarkers (23).
Appropriate biomarkers should be stable and non-invasive, and ought to be disease-specific for reliable and accurate measurement across a diseased population (24). It is recommended that due to the prevalence of miRNA regulation, it should participate in a wide range of human-specific diseases. It is proposed that such regulators have critical functions, such as oncogenes or tumor suppressor genes in various types of cancers (25). For instance, miR-16, as well as miR-15, frequently undergo deletion in different types of leukemia (26) and miR-182, miR-96, and miR-183 expressions correlate with the progression of non-small cell lung cancer (27). Also, miR-423-5P (28), miR-16 (29), miR-139-5P (30), miR-182, and miR-187 (31) are among the detected miRNAs used as biomarkers in cancer diagnosis. They are also involved in some other diseases, such as immunological, psychiatric, and neurodegenerative ones (32). Down-regulation of the biogenesis factors (33), a mutation in the miRNA locus (34), or epigenetic changes-e g, hypermethylation (35), can perturb the miRNA function. Prior to investigating the role of miRNA in a disease, it should be predicted and annotated, according to its specific expression pattern. Then, by artificially altering the expression level of miRNAs, the initiation and progression of the diseases could be controlled. This issue is used to treat cardiovascular diseases-eg, cardiopulmonary resuscitation (36), cardiac calcium signaling (37), and cardiac repair after myocardial infarction (38). Thus, predicting miRNA is a substantial step of primary analysis in the clinical context.
The discovery of novel miRNAs eventually results in an alteration in treatment attitudes, enhanced clinical results, higher allocation of health care resources, and increased utilization of miRNA-based therapy (39). More miRNAs are detected by extensive cloning and sequencing. The major limitation of miRNA detection by cloning is that it is troublesome to find miRNAs with low expression levels, expressed per cell at different stages of development, or exhibit tissue-restricted expression. Nevertheless, the process of miRNA cloning, according to their physical characteristics, such as post-translational modifications or nucleotide sequences, is not easily achieved. Additionally, expensive and time-consuming cloning techniques added more limitations as well (40). Computational algorithms can be used to provide quick, efficient, and inexpensive methods to detect and predict miRNA coding sequences in the genome. It should be confirmed in-vitro by examining the expression of the endogenous miRNA mature form (41).
Lai et al., according to the expression profiling and bioinformatics analyses, suggested about 24 new target genes for human miRNAs (42); furthermore, Hoballa et al., consistent to the bioinformatics prediction, introduced two novel miRZa-3p and miRZa-5p, which target SMAD3 and IGF1R genes (43).
Bentwich et al., also introduced a total of 89 novel human miRNAs in a broad study, combining bioinformatics predictions with microarray analysis and sequence-directed cloning (44). Dokanehiifard et al., using the SSCprofiler, UCSC genome browser, and several other databases, predicted and validated two novel miRNAs in the TrkC gene, as well as hsa-miR-6165 in the NGFR gene, and also investigated their possible association with colorectal cancer (45). Additionally, in a similar fashion, they predicted and confirmed a new miRNA in the PIK3KCA gene with a possible role in colorectal cancer (46).
Lim et al., using RNAfold web server to analyze folding and minimum free energy, predicted structures such as miRNA precursors and identified 38 novel human miRNAs (47); Wu et al., completely validated the novel hsa-miR-3675b that inhibited proliferation of human breast carcinoma cells (48).
5.1. Conclusion
The current study aimed at scanning the F8 gene in the contemplation of predicting and identifying potential sequences for the expression of mature miRNAs. Hemophilia A is a monogenic disorder; hence, a vast majority of the known and recorded human miRNAs affect the expression levels of their coding genes.
The present study used a highly accurate and reliable database. Hopefully, the proposed candidate sequences are experimentally approved in future studies and, subsequently, have a high impact upon initiation, advancement, and improvement of miRNA-based medicines aimed at treating and healing the long-suffering patients with hemophilia A.