1. Background
One of the multicellular organism hallmarks is cell-cell interaction guided by straight cell contacts or secreted molecular transition. Another intercellular communication mechanism identified over the last two decades is the intercellular transition of extracellular vesicles (EVs). Such tiny membrane vesicles carry signals to long distances from generating cells and influence numerous cell life aspects (1) Exosomes are membrane-derived nanovesicles with a size of 30 - 100 nanometer (nm). They are generated by all cell types from endosomes (2). Exosomes can serve as mediators of cell-cell communication by transmitting and sharing their charges and altering the biological properties of recipient cells (3, 4). Exosomes are secreted by various normal and cancer cells. Cancer cells develop more exosomes, which induce changes in recipient cells (near or distant from the tumor), including angiogenesis, invasion, metastasis, and resistance to chemotherapy (5, 6).
Exosomes derived from tumors become momentous agents that promote contact among the nearby cells and influence cell activities linked to cancer growth or development. A high-efficiency procedure for the isolation and characterization of exosomes generates crucial facts to better understand their specific role in cancer (7). Biological fluids and EVs can be obtained from cell lines in culture media. Strives made to take conventional approaches to EV isolation to understand their function and form (8, 9).
Despite the numerous reports published on comparative methods for EV isolation, no related guidelines for clinical practices are in hand yet. Differential centrifugation is among the most widely used EV isolation processes (10).
2. Objectives
Concerning many roles recently assigned to exosomes, the characterization of these nanostructures raised interest. The current study aimed at isolating exosomes derived from MDA-MB 231 cell culture media using ultracentrifugation and reporting the characteristics of these vesicles.
3. Methods
3.1. Cell Culture
The human breast cancer MDA-MB-231 cell line (in-vitro osteolytic form) was grown in Dulbecco’s Modified Eagle Medium (DMEM) augmented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution at 37°C in 5% humidity and 5% (v/v) CO2 (11).
3.2. Exosome Collection and Purification
MDA-MB-231 breast cancer cells in the 3rd passage were washed twice with phosphate-buffered saline (PBS) when covering 80% of the flask surface. Then, the cells were inoculated into a fresh cell medium with exosome-free FBS. After 24 - 48 hours, the culture was centrifuged to separate cellular residue from larger vesicles at 300 g for 10 minutes and 2000 g for 15 minutes, and then filtered on a 0.22-µm filter. Finally, ultracentrifugation at 100,000 g for 70 minutes was performed. The supernatant was discarded, the exosome pellet was suspended in 1 mL of PBS, and stored at -20°C (12, 13).
3.3. Transmission Electron Microscopy
The exosomes were sonicated for 30 minutes for homogenization. For this purpose, exosomes were fixed with 1 mL of 2.5% glutaraldehyde in 0.1 M sodium cacodylate solution for one hour at 4°C. Then, they were placed on 200 mesh copper grids, allowed to dry at room temperature, and exposed to 3% (w/v) uranyl acetate in water for 20 seconds. Then the grid was rinsed with PBS and allowed to dry at room temperature. Imaging was performed at 120 kV voltage (14).
3.4. Scanning Electron Microscopy
The suspension obtained by ultracentrifugation was sonicated. A small amount of sample was fixed with 2.5% paraformaldehyde and diluted in distilled water. The sample was dehydrated with ethanol, transferred to silicon chips, and placed in the acetone for one minute to be dehydrated and dry. Then it was washed with water and dried. The photos were taken by SEM (LMU TESCAN BRNO-Mira3) with the selected voltage of 30 kV (13).
3.5. Atomic Force Microscopy
The exosomes were sonicated for 30 minutes for homogenization. First, 3 µL of purified exosomes were fixed with 100 µL of 2% paraformaldehyde; then, a small drop of the solution was placed on a slide. After about 15 minutes, when the sample was dry, AFM (JPK-Nano Wizard II) was used to investigate (15, 16).
3.6. Dynamic Light Scattering
The exosomes were sonicated for 30 minutes for homogenization. For investigating the size of the exosomes with DLS, approximately 300 µL of the exosomal solution was inoculated into 1 mL PBS. The prepared sample was inserted into the device, and the results were analyzed. The DLS measurements were analyzed with a Zetasizer Nano (100Z-Horiba-SZ) (17).
4. Results
4.1. Characterization of Exosomes Derived From MDA-MB-231 Cells
4.1.1. Transmission Electron Microscopy
The morphology and size of the obtained exosomes were explored by TEM. The membrane integrity of exosomes was maintained during the isolation process so that their spherical structure remained intact. The particle sizes were within the reasonable range for spherical-shaped exosomes (30 - 100 nm) (Figure 1).
4.1.2. Atomic Force Microscopy
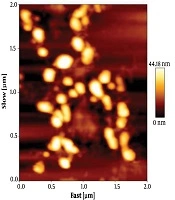
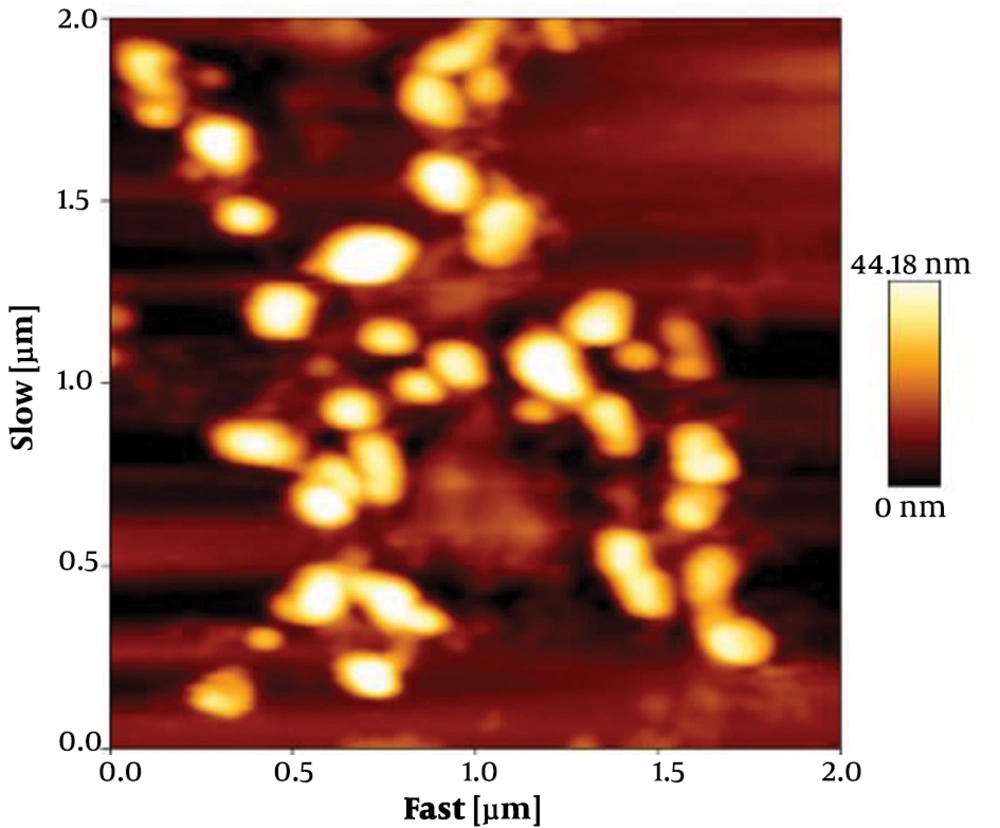
The size of exosomes was analyzed by tapping mode. Data analysis showed that the size of particles were within the acceptable range of 30 to 100 nm. The collected data showed that the membrane integrity of exosomes was maintained during the isolation (Figure 2).
4.1.3. Dynamic Light Scattering
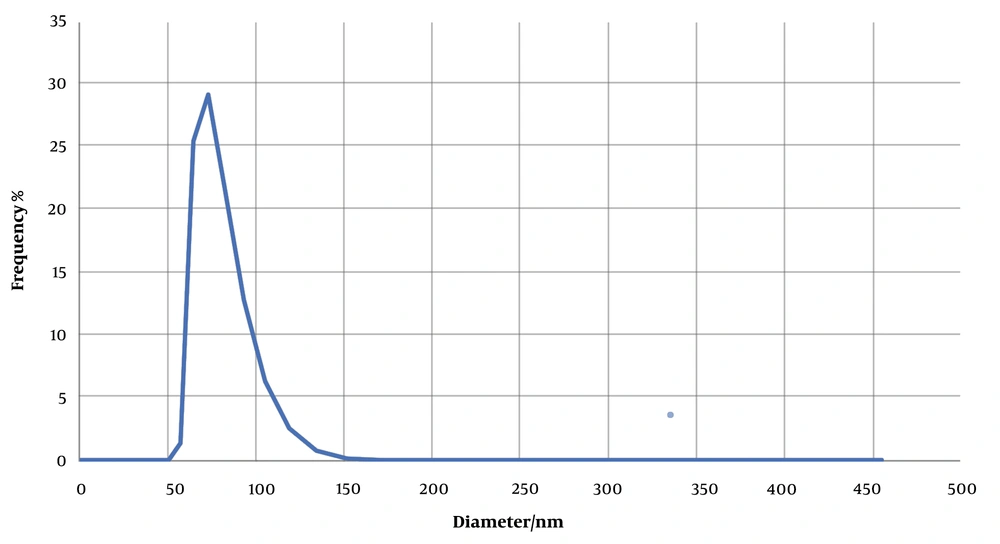
The size of exosomes isolated from MDA-MB-231cells was determined using a Zetasizer Nano and DLS technique. For analysis, 1 mL of the prepared sample was poured into a low-volume cuvette. Assessment of the distribution of sizes by DLS showed a peak of approximately 70 nm for the population of extracted exosomes (Figure 3).
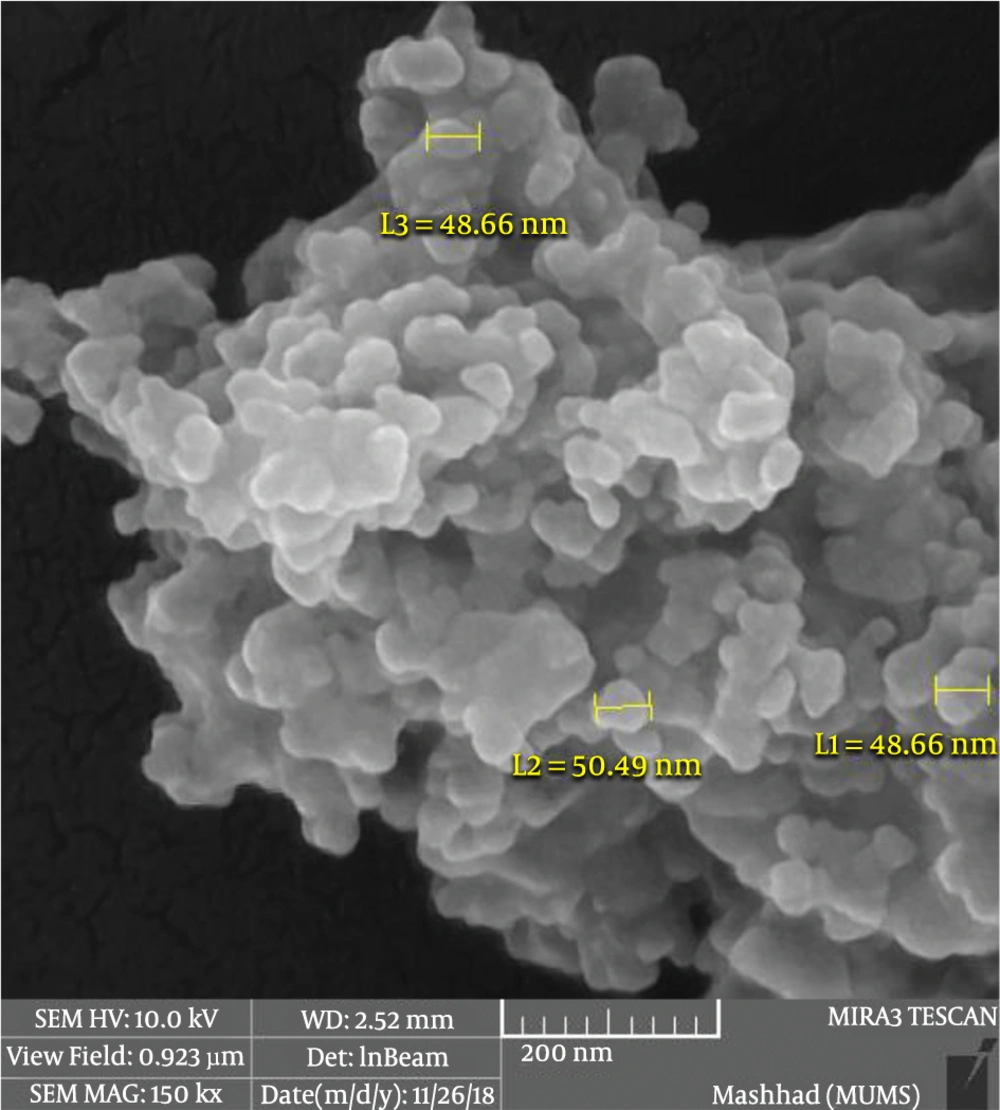
4.1.4. Scanning Electron Microscopy
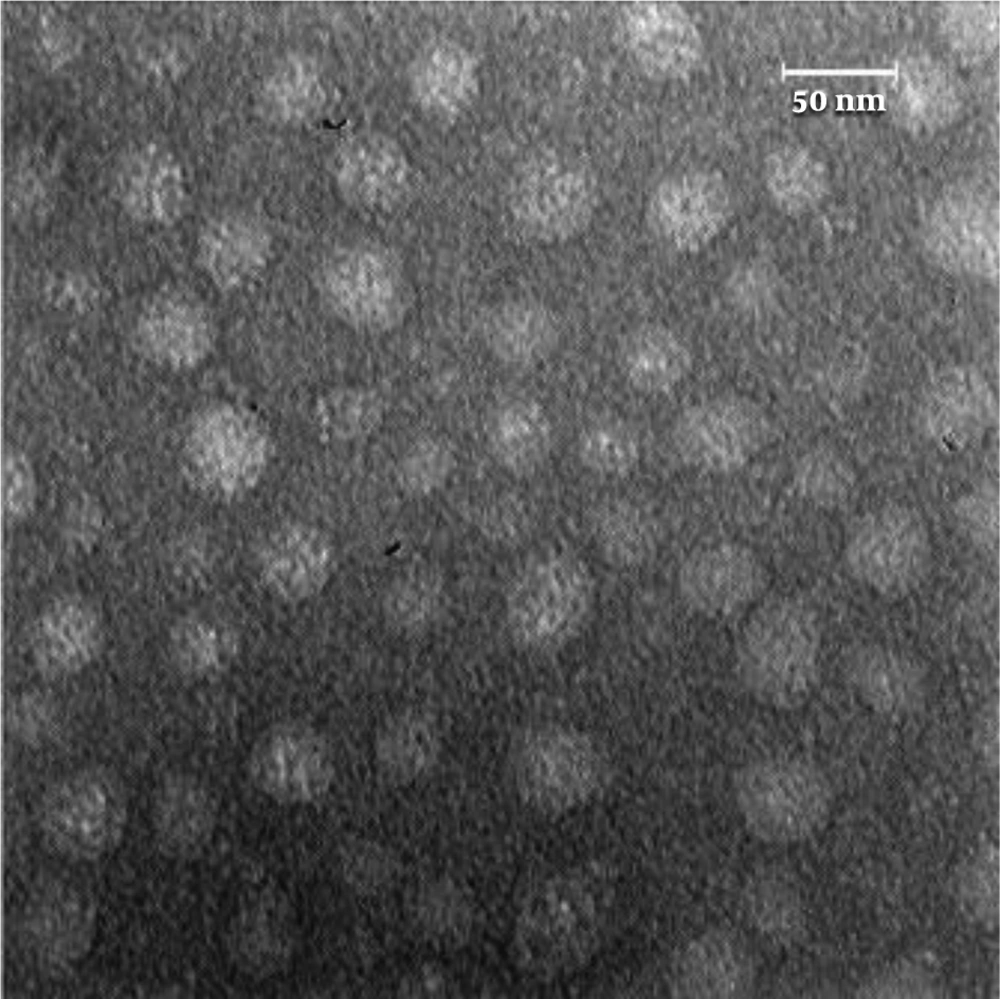
Exosomes were analyzed by SEM to confirm their morphology and size, as described in the Materials and Methods section. As shown in Figure 4, the exosomes were spherical, and their sizes ranged from 30 to 100 nm. The results confirmed the purity of exosomes isolated from the cell culture media (Figure 4).
The obtained results confirmed the methods employed for the isolation and characterization of exosomes.
5. Discussion
Under normal and pathological conditions, all cells release vesicles called exosomes, which attracted researchers’ attention in recent years due to their numerous roles (17). Exosomes contain genetic information and proteins, which vary across the cells they originated. Information can be transmitted through the exosome to the recipient cell, near or far from the release site, and induce functional and phenotypic changes in the target cell (18, 19). It opens up the possibility that cancer cells exert their biological effects by transmitting information to the target cells through exosomal transfer. Therefore, exosomes can act as a paracrine agent between cancer or tumor cells and normal cells (20). Exosomes derived from tumor cells are considered as agents promoting contact among nearby cells and influencing cell activities linked to the growth of cancer (7). As a result, the purification and identification of exosomes, as the first step towards human health promotion, is an important topic of discussion in basic research and clinical applications (20).
Hence, it is necessary to develop appropriate methods to isolate and characterize exosomes, which may be useful in the prognosis, prevention, and treatment of diseases. Therefore, due to the importance of the pure isolation of exosomes to understand their modes of action and application in medical sciences, the present study aimed at isolating exosomes derived fromMDA-MB-231 cells by differential centrifugation and filtration before ultracentrifugation to obtain pure exosomes and evaluate their size and appearance. The exosomes may be used as biomarkers for the early detection of breast cancer. To date, various strategies are proposed for exosome isolation. The difference in these methods is based on their efficiency (19). In standard isolation methods, the removal of excess particles and protein concentration are also considered (21). Previous researches reported ultracentrifugation as an efficient and inexpensive method, in which a considerable amount of exosomes precipitates, although it leads to exosome aggregation. Several studies suggested ultracentrifugation in sucrose gradients, which does not cause exosomal aggregation. However, the contamination of exosomes with sucrose, as a gradient reagent, may interfere with exosome functional studies (20). Thus, every method of purification has advantages and disadvantages, and it may be suitable to use a combination of the two. An effective manner due to using the differential centrifuge and filtering of the culture medium of the breast cancer cells was performed before ultracentrifugation. Then, exosomes were sonicated for homogenization and identified based on size and appearance using reliable methods. The current study findings showed that exosomes derived from MDA-MB-231 cells were spherical vesicles with 30 - 100 nm in diameter, indicating that the employed strategy was effective. The current study results were consistent with those of other research. Reimondi et al. (22) analyzed the size of myeloma-derived exosomes using the DLS method and reported that the U266- and OPM2-derived exosomes were approximately 100 and 50 nm in diameter, respectively. In another study, the size of mesenchymal stem cells isolated by ultracentrifugation, examined by DLS and TEM, were 80 and 40 - 100 nm, respectively.
In general, the technique proposed for isolation, verification, and identification of exosomes is advantageous in analyzing their roles, functions, and applications. However, there is no gold standard for exosome isolation. Further studies are required to achieve a uniform protocol for exosome isolation.