1. Background
Bile acid (BA) secretion is a critical biological process regulating cholesterol and lecithin excretion in the human body, and its disruption can result in cholestasis (1). Cholestasis affects the function of different organs due to the aggregation of bile in the body, including the male reproductive system (2). Previous studies have shown significant changes in reproductive organs’ weights, suppression of testosterone production, reduction of sperm count and motility, histopathological changes in testes, and increased testicular oxidative stress biomarkers in the testes of the rat models of cholestasis (3). The inflammatory reactions induced by cholestasis and the subsequent macrophages’ entry increase autophagy in spermatocytes and disrupt spermatogenesis (1).
Micro RNAs (miRNAs) account for 1-5% of the human genome and regulate at least 30% of protein-encoding genes (4). MiRNAs have important roles in regulating liver function and modulating its pathogenic conditions; for example, miR-let7a-5p affects the expression of adenosine triphosphate–binding cassette subfamily C member 2 (ABCC2/Abcc2) gene that is up-regulated in obstructive cholestasis (5). In humans, hepatic multidrug resistance protein 3 (MDR3), which is down-regulated in the cholestatic liver, is directly regulated by miR-378a-5p (6). Several lines of evidence have suggested significant roles for miRNAs in spermatogenesis and male fertility (7). They also regulate cellular responses to the hormones secreted by Sertoli cells (8). Because of their important regulatory roles, they can be considered as biomarkers for screening and identifying male infertility (9). MiR-34c is one of the significant miRNAs contributing to mammalian spermatogenesis (10, 11) and is highly expressed in adult male testicles where it controls the proliferation and differentiation of spermatogonia cells (12) and is involved in the four stages of spermatogenesis (13). Altered expression of miR-34c has been shown in various infertility disorders such as oligoasthenozoospermia, asthenozoospermia, and azoospermia (10, 14). Investigating the expression profile of miRNAs in azoospermic patients suggested miR34c to be essential for spermatocyte meiosis (15) and the development and formation of spermatocytes and sperms in adult primates (16). It was shown that increased miR-34c expression in mature mouse testes contributed to sperm production and the regulation of mouse embryonic stem cells’ differentiation to male germ cells (11). In one study, the over-expression of miR-34c led to the up-regulation of the genes associated with meiosis, including Nanos3, Scp3, and Stra8, during mouse spermatogenesis (17). The computational predictions illustrated on http://mirgate.bioinfo.cnio.es/miRGate/ revealed that mir-34c-5p targets THY-1, Fibroblast Growth Factor 2 (FGF-2), and Caspase-3 (CASP-3) genes (Table 1). Fibroblast Growth Factor 2 is secreted from various cells in the testis, and it has been revealed that FGF-2-deficient mice were unable to produce normal spermatozoa (18). Besides, THY-1 or CD90 is a spermatogonia stem cell factor that in rodents indicates the presence of SSCs in testes (19). Finally, CASP-3 encodes a protease involved in cell death, and its activation under cholestasis-induced oxidative stress promotes cell apoptosis in the testis (20).
| Input Genes | Transcripts | miRNA | Start | Stop | Computational Predictions |
|---|---|---|---|---|---|
| Thy-1 | ENSRNOT00000008685 | rno-mir-34c-5p | 662 | 684 | 3 |
| Casp-3 | ENSRNOT00000014095 | rno-mir-34c-5p | 159 | 184 | 1 |
| Fgf-2 | ENSRNOT00000023388 | rno-mir-34c-5p | 10 | 18 | 2 |
The Computational Predictions of miR-34c-5p on Thy-1, Casp-3, and Fgf2 Genes
2. Objectives
Regarding the importance of mir-34c for proper spermatogenesis and the relationship between cholestasis and testicular dysfunction, in the current study, we aimed to study the expression of miR-34c-5p and its target genes (i.e., THY-1, FGF-2, and CASP-3) in an animal model of cholestasis.
3. Methods
3.1. Cholestasis Induction in Animals and Tissue Collection
In this study, male Rattus norvegicus Wistar rats of 2 - 2.5-month-old, weighing between 200 and 300 g were transferred to our laboratory and kept in a controlled environment. Rats were divided into two groups; the control and cholestasis model (n = 6 per group). In order to create cholestasis models, rats were anesthetized by the intraperitoneal injection of ketamine and xylazine. Then by making an incision in the abdomen’s midline, the common bile duct was exposed, ligated close to the liver hilum just beneath the bifurcation, and then dissected (21). The rats were placed in cages and housed for four weeks in controlled conditions. After four weeks, cholestasis rats were sacrificed by CO2 inhalation, and their testicles were collected for further analyses.
3.2. Histological Assay
Paraffin-embedded specimens of testes were prepared following ethanol dehydration and xylene clearance. The tissues were sectioned at the thickness of 7 µm, and the sections were deparaffinized in xylene and rehydrated by ethanol. Then tissue slides were embedded into hematoxylin for two minutes. For eosin staining, the sections were dehydrated using ethanol and then kept in eosin for two minutes. Finally, the sections were cleared using xyleneand the slides were examined under a light microscope
3.3. Evaluation of ALP, AST, and ALT Liver Enzymes
In order to confirm the creation of cholestasis, serum levels of alkaline phosphatase (ALP), aspartate aminotransferase (AST), and alanine aminotransferase (ALT) enzymes were assessed in the experimental rat models. Briefly, four weeks after cholestasis induction, blood was collected from the heart, and after 30 minutes of staying at room temperature and blood clot formation, the serum was separated and assayed for the mentioned enzymes using special kits (Pars Azmoon, Iran).
3.4. Real-Time PCR
The gene expressions of THY-1, FGF-2, CASP-3, and miR-34c-5p were analyzed in the testicular tissues of the control and cholestasis groups using Real-Time PCR. In summary, RNA was extracted from testicular tissues according to the manufacturer’s protocols (QIAGEN, Germany). The purity and concentration of RNA were assessed using the Agilent 2100 bio-analyzer (Agilent Technologies, USA). The synthesis of cDNA was carried out by the Revert Aid First-Strand cDNA Synthesis Kit (Fermentas, USA) according to the manufacturer's recommendations. Real-Time PCR was conducted using the ABI 7500 (7500 Real-Time PCR Software) instrument and SYBR Green Master Mix (QIAGEN, Germany) in a final volume of 20 µL. The thermal cycling was performed based on the following program; initial denaturation at 95°C for 15 minutes, followed by 40 cycles of annealing at 58°C for 10 seconds and amplification at 72°C for 15 seconds with a final extension at 72°C for 10 min. The Gapdh gene was utilized as an internal control, and the fold-change of the genes was calculated by the 2-ΔΔCT method. Primers have been listed in Table 2.
| Genes | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Gapdh | AAGTTCAACGGCACAGTCAAGG | CATACTCAGCACCAGCATCACC |
| Fgf-2 | AGAAGAGAGAGGAGTTGTG | GGTAAGTGTTGTAGTTATTGGA |
| Casp-3 | AAGTGATGGAGATGAAGGAGT | CAGGCGTGAATGATGAAGAGT |
| Thy-1 | GATATGGATGAGGGAAGTTGG | ATAGAAGGGGGCTGAGAATGA |
| U6 | TGCTTCGGCAGCACATATAC | AGGGGCCATGCTAATCTTCT |
| miR-34c-5p | AGGCAGTGTAGTTAGC | GTGCAGGGTCCGAGGT |
Sequences of the Primers Designed for This Study
3.5. Statistical Analysis
All experiments were conducted at least three times. The difference between the groups was evaluated by one-way ANOVA and Tukey post hoc test. The level of statistical significance was considered at a p-value of less than 0.05.
3.6. Ethical Statement
All experimental procedures in this study were approved by the ethical committee of Kharazmi University under the code of 20288.
4. Results
4.1. Tissue Structure of Testes in Cholestasis Rats
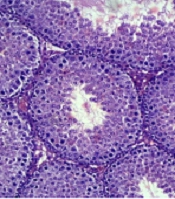
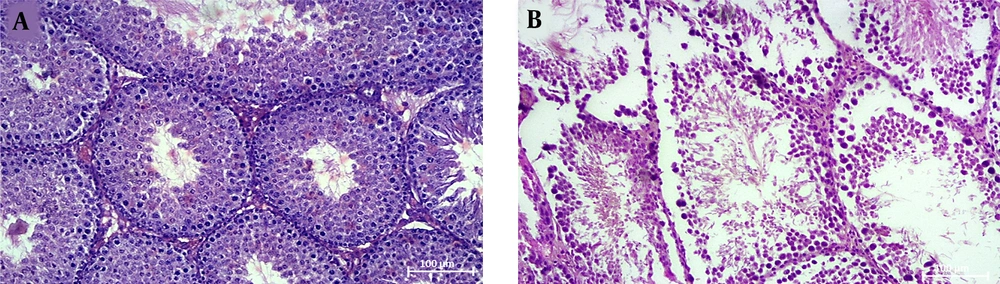
A comparison of seminiferous tubular structure between healthy and cholestasis mice showed that cholestasis induction damaged seminiferous tubular structure, characterized by displaced cells, the presence of intracellular vacuoles, and disrupted tubular basement membranes in some parts (Figure 1).
4.2. Liver Enzymes
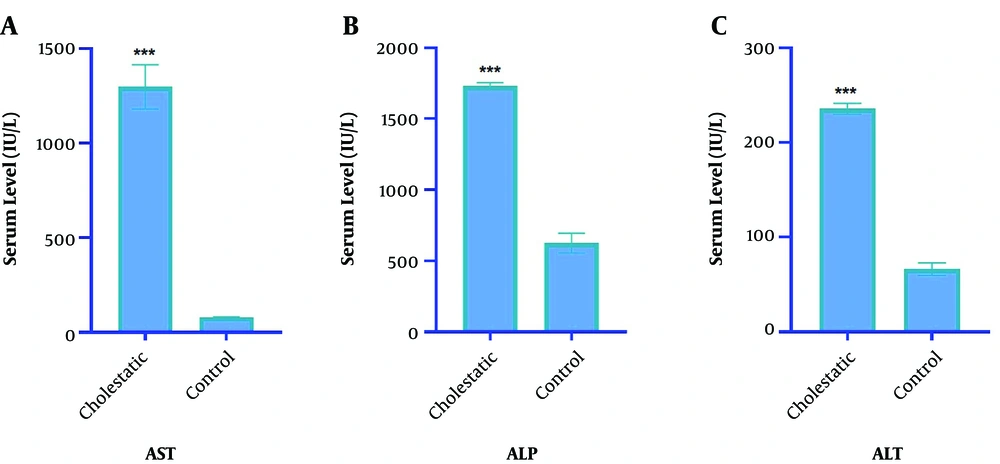
The results showed that ALP, ALT, and AST liver enzymes significantly increased in the cholestasis group compared to the control group (P < 0.001), revealing a rise up to 17 folds four weeks after the surgery. These results indicated that cholestasis was successfully induced in animals (Figure 2).
4.3. MiR-34c-5p Expression
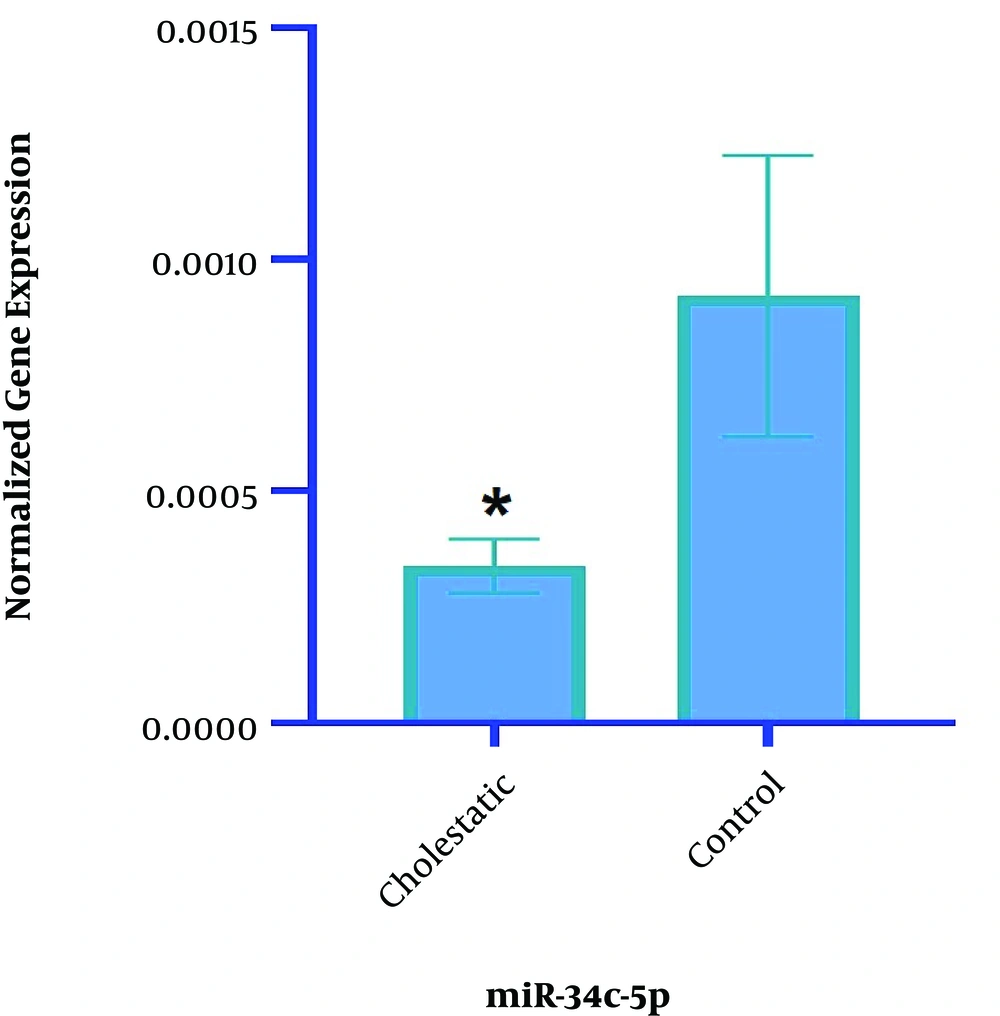
The results of Real-Time PCR showed that the expression of miR-34c-5p significantly decreased in the cholestasis group compared with the control group (2-1/1125, P = 0.05) (Figure 3).
4.4. The Expression of MiR-34c-5p Target Genes
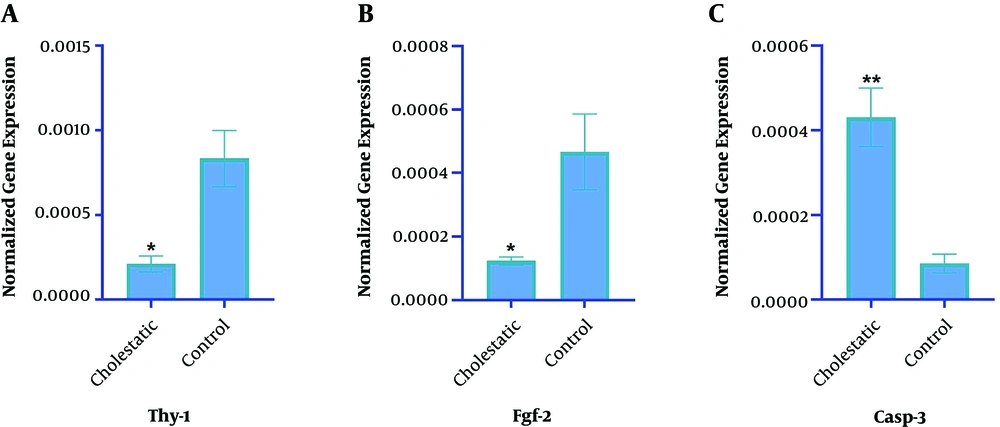
The impact of altered miR-34c-5p expression was assessed on the expressions of THY-1, FGF-2, and CASP-3, as its target genes, in both the control and cholestasis groups (Figure 4). The results showed that the expressions of THY-1 (2-2/0525, P = 0.01) and FGF-2 (2-1/7915, P = 0.02) markedly reduced in the cholestasis group compared with the control group. Inversely, the expression of the CASP-3 gene considerably increased in the cholestasis group compared with the control group (2+2/43, P = 0.00). Respective melting curves have been shown in Figure 5.
5. Discussion
Cholestasis causes the accumulation of BAs in the serum, and excessive amounts of BAs induce apoptosis in spermatids and germ cells through activating the Farnesoid X receptor alpha (FXRα) (22). In this context, similar to young mice, fxrα−/− male mice demonstrated a greater number of undifferentiated germ cells and fertility capacity (23). In the present study, to better understand the effects of cholestasis on testes, we examined the expression of miR-34c as one of the critical miRNAs involved in spermatogenesis. Our results indicated that the expression of miR-34c in the testicles of cholestasis rats decreased compared to controls. Several pieces of evidence, especially the role of inflammation, may explain this finding. One of the adverse effects of cholestasis on testes is infiltration by macrophages, subsequently leading to inflammation. Shi et al. demonstrated that after bile duct ligation, the number of cells expressing proliferating cells nuclear antigen (PCNA+) significantly reduced while the expressions of apoptosis-related genes such as CASP-3 and Bcl-2-associated X protein (BAX) elevated (24). Our finding also showed a higher expression of CASP-3, which could boost cellular death in testes of cholestasis rats. Two studies have shown that inflammation changes miR-34c expression in testes; the first study showed that six hours after receiving an intraperitoneal injection of an inflammation-inducing agent, bacterial lipopolysaccharide (LPS), the expressions of miRNAs, including miR-34c, gradually reduced in the testes of mice (25). The other study showed that miR34c-5p was significantly diminished in the testicular tissues of patients with cryptorchidism, as well as in murine models of cryptorchidism (26). In this regard, a previous study highlighted the presence of an inflammatory state in cryptorchid testes (27).
Our findings also demonstrated that cholestasis induction was accompanied by decreased THY-1 and FGF-2 expressions, supporting the hypothesis that miR-34c-5p had a regulatory site on the mentioned genes and the fact that altered miR-34c level might change THY-1 and FGF-2 expressions. The downregulation of THY-1 and FGF-2 genes could disrupt spermatogenesis in cholestasis animals. THY-1 is a marker for undifferentiated spermatogonia cells (28). A study performed by Ryu et al. showed that all spermatogonia stem cells in rats’ testicles expressed the THY-1 gene (29), and another study conducted by Yao et al. revealed that 90% of human spermatogonia were positive for this marker (30). A comparison between THY-1+ spermatogonia cells and THY-1− somatic cells revealed that miR-34c was highly expressed in the THY-1+ cell population (31). Likewise, FGF-2 encodes a growth factor secreted from various cell types in the testis (32) and is essential for normal spermatogenesis and sperm motility. It was noted that FGF-2-deficient mice were unable to produce normal spermatozoa (33). This factor is also necessary for the proliferation and differentiation of spermatogonia cells (34).
5.1. Conclusions
In conclusion, the current study revealed that cholestasis might affect the function of testes and spermatogenesis via dysregulating the expression of miR-34c. Considering the importance of miR-34c in several steps of sperm production, its dysregulation can be associated with the impaired expression of the genes needed for the development and viability of spermatocytes, such as THY-1 and FGF-2.