1. Background
Acinetobacter baumannii (A. baumannii) is an opportunistic pathogen that has always been identified as one of the most important pathogens of nosocomial infections (1). One of the major problems in treating and preventing infections caused by A. baumannii is its antibiotic resistance (2). Acinetobacter baumannii develops multidrug resistance, which has been demonstrated by a variety of antibiotic resistance mechanisms, including the activation of efflux pumps, production of beta-lactamase enzymes, reduction of foreign membrane permeability, and synthesis of enzymes such as phosphoryl transferases and acetyltransferases that cause resistance to aminoglycosides (3).
In addition to the above, there is another mechanism in the antibiotic resistance of A. baumannii called biofilm (4). Biofilms are complex bacterial communities attached to surfaces that are created by an extracellular matrix produced by bacteria. This matrix is composed of polysaccharides, DNA, and proteins. Bacterial biofilms cause many problems in medical, industrial, and environmental fields. Adherence of bacteria to surfaces is an important issue in ecology, biotechnology, bio-pollution, and wastewater treatment. Various proteins are involved in the development of this resistance, including proteins that create and strengthen the biofilm structure (5). The first member of this group of proteins, called BAP, has been identified in the bacterium A. baumannii (6). These proteins are located on the outer surface of bacteria. They consist of a central nucleus consisting of iterations of similar sequences. Bacteria are able to form biofilms, thus playing an important role in infectious pathogens. The structural features of these proteins include their presence on the outer surface of bacteria, high molecular weight, and the presence of a central nucleus consisting of repetitive sequences of similar sequences. These proteins give the bacterium the ability to form biofilms, and thus, play an important role in infectious pathogens (7).
Today, researchers are looking for new solutions to treat drug-resistant bacteria (8). One area of application, in this case, is the use of newly synthesized drug structures in the treatment of microbial infections. So far, numerous studies have shown that various compounds of oxadiazole derivatives have antibacterial properties against A. baumannii strains (9). Among these compounds, we can mention fluorophenyl compounds with thiol (10), fluorophenyl with chlorophenyl group (11), etc. Due to the antibiotic resistance of A. baumannii, few classes of antibiotics can be used to treat infections caused by this bacterium.
In this study, the effect of the synthesized materials was investigated for the first time on the expression of biofilm-inducing Bap gene in A. baumannii isolates. The effect of synthesized derivatives on the bacteria present in the biofilm, especially in deeper layers, was comparable to antibiotic treatment and phagocytic function of immune cells through inhibiting the formation of biofilm and regulating the development of the degenerative environment. Just as it is important to identify genes involved in the biofilm formation and pathogenesis of this bacterium, it is also important to recognize the environmental factors playing a role in regulating the expression of these genes (12).
2. Objectives
The aim of this study was to investigate the impact of new derivatives of 1, 3, 4-oxadiazole based on the 1, 3, 4-oxadiazole-2-yl-piridin-2-yl-methanol group on the expression of A. baumannii Bap gene, which is effective in promoting the biofilm formation ability of drug-resistant specimens.
3. Methods
This study was conducted in the genetic laboratory of Islamic Azad University, Tehran branch, with code number 10130553971011 in 2019. Starting materials, solvents, and culture environments were obtained from Merck, Germany, and used moving forward without any more filtration.
3.1. Preparation of Samples
In this study, 100 urine samples suspected of Acinetobacter infection were cultured on plates (Mueller Hinton agar). The samples were from patients with severe urinary tract infections and were delivered from the research department of Shariati and Sarem hospitals in Tehran. In the microbiology laboratory of Islamic Azad University, Central Tehran Branch, by using biochemical methods, 10 strains of A. baumannii were identified. A pure culture of all isolates of A. baumannii strains was prepared on blood agar medium, and then the cultures were incubated for 24 h at 37°C. To preserve the bacteria, they were inoculated in liquid culture medium containing glycerol and then stored at -70°C. The presence of Bap gene was confirmed by the PCR technique. In addition to clinical specimens, the standard strain and reference A. baumannii ATCC25923 were used, which makes the test conditions under quality control. The standard strain was obtained from the Iranian Research Organization for Science and Technology (IROST).
3.2. PCR to Confirm the Bap Gene
The presence of Bap gene was confirmed by molecular PCR using CinnaGen fermentase kit. The reaction mixture for PCR of the Bap gene was as follows: (1) MasterMix (1x): 12.5 µL; (2) primer F (0.1 - 1 µM) 1 µ; (3) primer R (0.1 - 1 µM): 1µ; (4) template DNA: 5 µ (20 pg), (5) sterile deionized water: 5.5 µ; the final volume was 25 µL, and PCR was set up to detect the Bap gene according to the following method. Initial denaturation: 95°C, 120 s, 1 cycle, denaturation: 93°C, 45 s, 35 cycles, annealing: 57°C, 60 s, 35 cycles, extension: 72°C, 30 s, 35 cycles, and final extension: 72°C, 90 s, 1 cycle. The primers used for the Bap and 16srRNA genes are listed in Table 1. Finally, six strains of A. baumannii with the Bap gene were selected for molecular studies.
| Gene Name | Primers | Product Size |
|---|---|---|
| BAP | Forward: 5'- TAGACGCAATGGATAACG -3' | 127 bp |
| Reverse: 5'- TTAGAACCGATAACGATACC -3' | ||
| 16s rRNA | Forward: 5'- TATCAGGACCATCTGGAGTAGG -3' | 110 bp |
| Reverse: 5'- CATCAACTTCACCTTCACGC -3' |
3.3. Synthesis of Derivatives
1, 3, 4-oxadiazole compounds were synthesized using a single-stage, high-yield method. Single-stage: (1) N‑iso-cyan-imino-triphenyl-phosphoran (1 mmol) + 2‑pyridine-carbaldehyde (1 mmoL) were dissolved in CH3COCH3 (7 mL); (2) in the next step, the carboxylic acid (1 mmoL) in CH3COCH3 (10 mL) was added to the previous solution. The final solution was stirred for 12 h by a magnetic stirrer at room temperature. The solvent was removed by evaporation, and the viscous residue was purified by flash column chromatography [silica gel powder: petroleum ether-ethyl acetate (4: 1)] (13).
3.4. Preparation of Derivatives Suspension
To prepare the stock solution, 1 g of each synthetic material was dissolved in 100 mL of dimethyl sulfoxide (99% DMSO) to prepare 0.1 mg/mL of each compound.
3.5. Broth Microdilution Test to Determine the Minimum Inhibitory Concentration (MIC)
To determine the MIC, the serial dilution method was performed in three replications in the culture medium. Thus, different dilutions from 1000 to 31.25 μL/mL of synthetic materials were added to 96-well plate wells. Then, 100 μL of microbial suspension was added to each well, and the plates were treated overnight at 37°C in a greenhouse. Finally, the turbidity of all wells was visually investigated and reported as MIC in each sample.
3.6. Treatment of Acinetobacter baumannii Samples with Synthesized Derivatives
To investigate the effects of synthesized derivatives (4a-4i) on the expression of the desired genes, A. baumannii samples were treated with the treated compounds, and then RNA extraction was performed. To do this, samples of A. baumannii, equivalent to half a McFarland, were cultured in a test tube containing 5 mL of broth nutrient medium. The MIC equivalent of the compounds was then added to each tube. After mixing, it was incubated for 42 h at 39°C. After incubation, the samples were used to extract RNA.
3.7. RNA Extraction
To extract RNA from CinnaPure-RNA kit with No. cat PR891620 of Cinnagen Company was used. Then, the accuracy of RNAs was measured by a NanoDrop, and the samples were placed at -80°C.
3.8. Total RNA Treatment of Acinetobacter baumannii Samples Before and After Proximity with Derivatives by DNase Enzyme
To convert RNA to cDNA and perform real-time PCR, RNA samples must be free of genomic DNA contamination; thus, before cDNA synthesis, all RNA samples were treated with DNase enzyme. RNA treatment was performed using the Fermentas kit during a two-step process. Then, 1 µg of RNA was added to a sterile nuclease-free microtube, and 1 µL of reaction buffer 10x DNase1 and 1 µL of RNase-free were added to the microtubes, and the microtubes were incubated for 30 min at 39°C. Finally, 1 µL of EDTA (25 mM) was added to each microtube, and the microtubes were placed at 65°C for 10 min. RNA concentration of the extracted samples was measured after treatment with DNase by a nanodrop device. After confirming the accuracy of RNA and removing the extra DNA, all RNA samples were quickly converted to cDNA, and its conversion was confirmed by electrophoresis.
3.9. Investigation of Bap Gene Expression
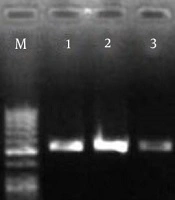
RNA samples of pathogenic strains were prepared immediately after quality assay with a nanodrop device. The accuracy of the cDNA reaction was confirmed by PCR on the agarose gel. The result of electrophoresis of the PCR product of cDNA samples is shown in Figure 1.
3.10. cDNA Synthesis
To make cDNA, Cinnagen Company RT PCR kit with the product code RTPL12 was used. The samples were then used for real-time PCR.
3.11. Real-time PCR
Real-time PCR technique was used to determine the expression of Bap and 16srRNA genes. The efficiency and melting temperature (TM) of these primers were evaluated and confirmed by ID Allele software. Requirements for real-time PCR were performed according to the protocol of Fermentase SYBR Green MasterMix kit and according to Table 2 in 36-house plates for Corbett machine. The amplification fragment contains 127 nucleotides for the Bap gene and 110 nucleotides for the 16srRNA gene. The annealing temperature was 57°C for the Bap gene and 51.8°C for 16srRNA gene. The preparation of real-time PCR samples and the conditions for real-time PCR are shown in Table 2. It should be noted that the real-time PCR was performed according to the MIC results and for 4i combination.
| Material | Final Concentration | Volume | |||
|---|---|---|---|---|---|
| qPCR SYBR green master mix | 1x | 12.5 µL | |||
| ROX dye 50x | 1x | 0.05 µL | |||
| F primer | (0.1 - 1 µM) | 1 µL | |||
| R primer | (0.1 - 1 µM) | 1 µL | |||
| Template | 1 µg | 1 µL | |||
| D.W | - | 9.45 µL | |||
| Total volume | - | 25 µL | |||
| Initial Activation | Denaturation | Annealing | Extension | Repeat | Final Extension |
| 95°С | 95°С | - | 72°С | 40 cycle | 72°С |
| 10 min | 20 sec | 20 sec | 20 sec | - | 1 min |
3.12. Quantitative Determination of Bap Gene Expression Before and After Exposure with a Concentration of 1 Mg/Ml of 4a, 4d, and 4i Derivatives Using Real-time PCR Technique
After real-time PCR, relative quantification can be performed in several ways, including using the standard curve of one gene and using the standard curve of both genes and the ΔCT method. The default of the ΔCT method is the relative parity of the cDNA amplification reaction efficiency with these two primers. Due to the equality of amplification efficiency of the two primers in the present study, the relative quantification of the cDNA of the studied genes in comparison with the cDNA of the 16srRNA gene (internal reference gene) was performed by the ΔCT method.
4. Results
4.1. Isolation and Diagnosis of Isolates from Clinical Samples
Four isolates were identified as A. baumannii using diagnostic and confirmatory microbiological/biochemical tests.
4.2. PCR to Confirm the Bap Gene
The results of electrophoresis of the Bap gene PCR products on agarose gel are shown in Figure 1.
4.3. Preparation of Derivatives
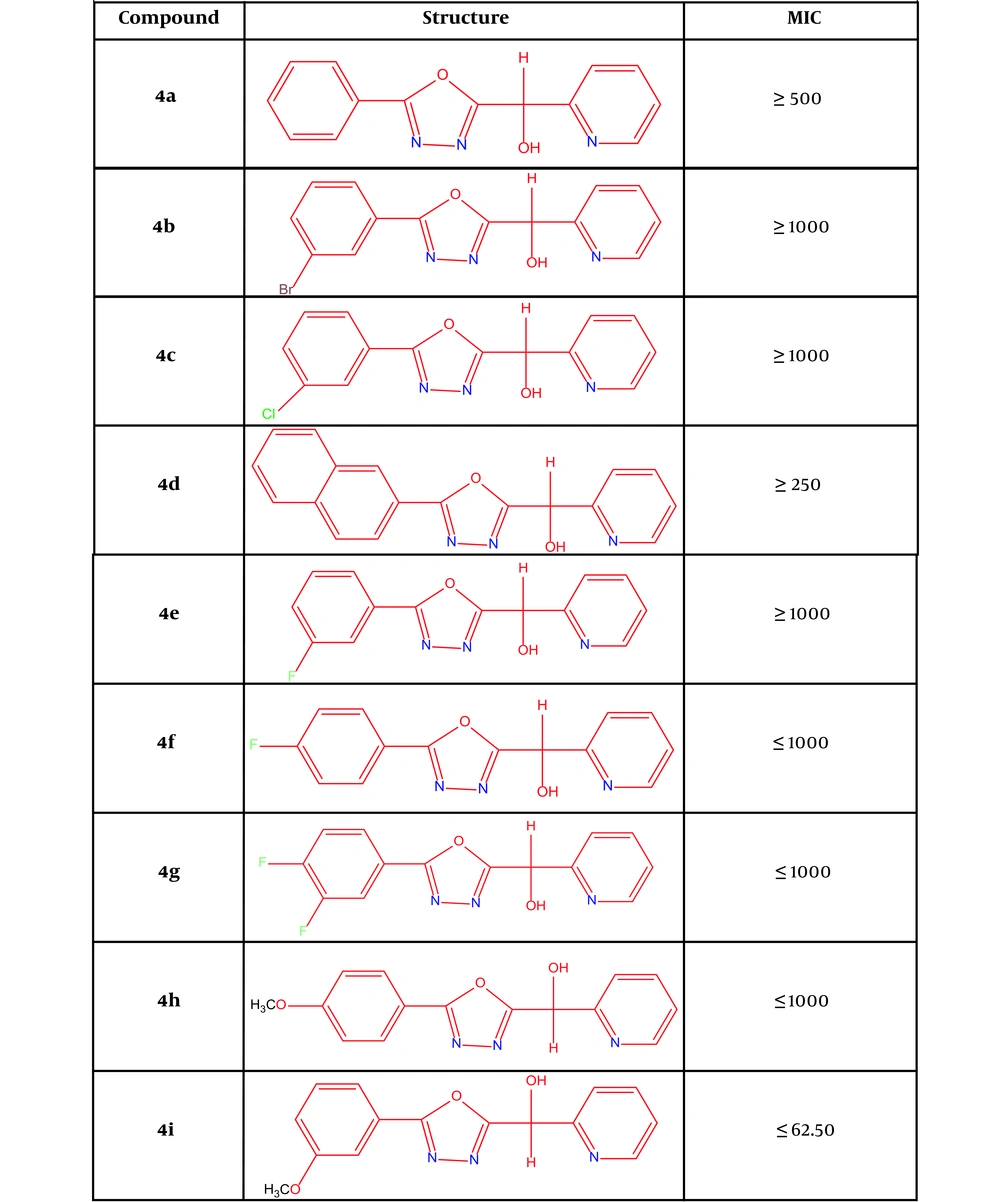
4a: (5-(phenyl)-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4b: (5-(3-(bromophenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4c: (5-(3-(chlorophenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol, 4d: (5-((naphthalene)-2-il)-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4e: (5-(3-(fluorophenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4f: (5-(4-(fluorophenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4g: (5-(3,4-(difluorophenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4h: (5-(4-(methoxyphenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol; 4i: (5-(3-(methoxyphenyl))-1,3,4-oxadiazole-2-il)(pyridine-2-il)methanol (Figure 2).
Structures of new derivatives of 1, 3, 4-oxadiazole (14) and MIC results
4.4. MIC Results
The results showed that the combinations of 4a, 4d, and 4i, respectively, have the minimum inhibitory concentrations of 500, 250, and 62.50. In this regard, compound 4i has the highest inhibitory effect among the synthesized compounds. Among the compounds, this compound (4i) was used to continue experiments (Figure 2).
4.5. Real-time PCR
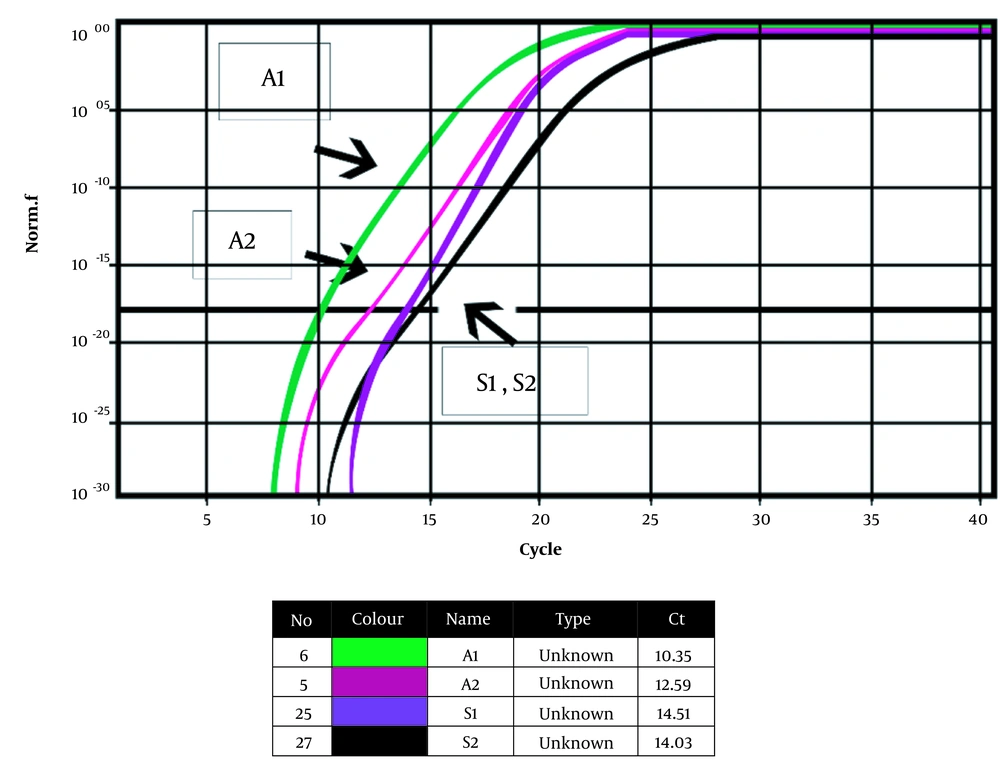
Figure 3 shows the Bap gene amplification diagram before and after treatment with compound 4i and the 16srRNA gene progression diagram as the housekeeping gene, along with a table of calculated CT values. A1 shows the Bap gene before treatment with compound 4i, A2 exhibits the Bap gene after treatment with compound 4i, S1 displays the housekeeping gene before treatment with compound 4i, and S2 shows the housekeeping gene after treatment with compound 4i. Controls template No (NTCS) did not have a reaction progression graph, but the samples had an exponential multiplication graph, and multiplication was performed. The 16srRNA gene was obtained before and after treatment at 14.51 and 14.03, respectively, considering that the difference in Ct before and after treatment with the combination of 4i was as small as 0.48; thus, the 16srRNA gene was used as a home gene and an internal standard. The results of the melting curve of the Bap gene in the clinical strain of A .baumannii treated with 4i showed that the main peak for the Bap gene occurs at 79 to 80°C. The ΔCt results of the Bap gene in the presence of the 4i combination are given below. The PFAFFL method was used to determine the expression of the Bap gene. In this method, it is assumed that the sample yield and internal control are equal to 100%, and the 2-ΔΔCt formula was used to investigate gene expression.
CT = 24.6 = 24.25
As can be noted, the combination of 4i reduced the expression of the Bap gene by about 24 times, but it had no effect on the expression of the 16srRNA housekeeping gene.
5. Discussion
Acinetobacter baumannii is the most common human pathogen in the genus Acinetobacter and is considered as one of the most important causes of nosocomial infections. One of the major problems in the treatment and prevention of infections caused by A. baumannii is antibiotic resistance, and carbapenems are the last line of treatment in infections caused by Gram-negative bacteria, including A. baumannii. The main factor of resistance can be biofilm production. There have been many studies on the resistance of Acinetobacter, the results of which have varied according to time and place (14-17). Therefore, one of the central goals of this research was to synthetize new structures for the treatment and elimination of this bacterium.
In recent years, the resistance and stability of bacteria to antibiotics has been a good reason to research the gene expressing biofilm (Bap) in various bacteria. For example, De Gregorio et al. conducted a study on proteins related to Acinetobacter biofilm. They stated that a large protein called Bap was involved in the formation of biofilms and the attachment of A. baumannii to host cells. The results of this group showed that several proteins with Ig-like repeats appear to be involved in biofilm formation (18). It can be said that several researchers have been interested in studying the resistance of A. baumannii to various antibiotics.
In 2016, Qi et al. conducted a study on the relationship between antibiotic resistance, biofilm formation, and biofilm specific resistance in A. baumannii. In their study, the relationship between antibiotic resistance, biofilm formation, and biofilm specific resistance in clinical isolates of A. baumannii was investigated (19). Other researchers have tried to show the effect of biofilm on the stability and survival of bacteria in the environment, especially in hospitals, which is a factor in causing nosocomial infections. Regarding biofilm structure, Fernández-Cuenca et al. demonstrated that in A. baumannii, adhesive cells that are coated with various coatings form an intermediate biofilm structure in the absence of nutrients. The findings indicate a rapid increase in the number of biofilm cells and a relative increase in the number of cells containing resistance (20).
This study proved that the presence of a biofilm in A. baumannii protects this microorganism against various antibacterial factors. The extraordinary ability of numerous structures containing the 1, 3, 4-oxadiazole ring as an antimicrobial agent has led to the use of derivatives of these structures in several studies. Various studies have shown that structures containing 1, 3, 4-oxadiazole have a significant ability to inhibit bacterial infectious diseases. In our previous study (14), we tested synthesis derivatives at a concentration of 1 mg/mL on a standard sample of A. baumannii, but in this study, despite a concentration 10 times lower and the use of a clinical sample, amazing results were obtained.
The 4i derivative in the presence of methoxyphenyl attached to the main structure had acceptable anti-biofilm properties against A. baumannii. Methoxyphenyl is one of the most effective compounds in various structures of 1, 3, 4-oxadiazoles, whose antibacterial and anti-cancer properties have already been proven. This compound (methoxyphenyl) has the ability to affect the cytoplasmic membrane, electron transport chain, metabolic activity, and gene synthesis and inhibit protein synthesis. By studying these compounds on other virulence genes of A. baumannii, we can study these compounds and other derivatives of these structures in laboratory and animal studies as a treatment or adjunctive therapy in the form of tablets or additives used in patients' food as well as hand and environment disinfectants to prevent the spread of infection caused by this bacterium in hospital settings.