1. Background
Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are opportunistic Gram-negative bacilli of the Enterobacteriaceae family and cause nosocomial infections. K. pneumoniae causes pneumonia, sepsis, and urinary tract infections, and E. coli is an indicator of urinary tract infection and causes pneumonia, sepsis, and bacteremia (1, 2).
Bacteria are immune to the harmful effects of antibiotics using a variety of mechanisms, one of which is the production of broad-spectrum beta-lactamases (ESBL) enzymes (3). Today, the prevalence of broad-spectrum beta-lactamase-producing bacteria is increasing and has raised concerns in treatment systems because they increase treatment costs, increase treatment duration, and increase mortality in patients (4). According to the results of studies, Gram-negative bacteria of E. coli and K. pneumoniae have the highest resistance, and mainly these strains carry ESBLs (5, 6). ESBLs are divided into 4 classes (A, B, C, D), which are mainly inhibited by inhibitors such as clavulanic acid, tazobactam, and sulbactam (7). According to this classification, the enzymes SHV, TEM, and CTX-M are in class A (8). More than 150 ESBLs have been reported so far. Broad-spectrum beta-lactamases are broad-spectrum hydrolyzing enzymes of cephalosporins (9). ESBLs are located on the plasmid, so they are easily transferable between different strains. The most common ESBLs are derived from TEM and SHV (10). Beta-lactamase TEM-1 is the most common beta-lactamase in Gram-negative bacilli Enterobacteriaceae family. Different types of beta-lactamase TEM have been formed by substituting amino acids for the active site of the TEM-1 enzyme, based on which about 130 beta-lactamase TEM has been identified. SHV beta-lactamase was first discovered by Pitton in 1972, which is a class A beta-lactamase. In the last two decades, widespread prevalence of CTX-M beta-lactamases has been reported in various regions (11-13). The first reports of CTX-M were recorded in the early 1990s. Unlike TEM and SHV, CTX-M beta-lactamases have a more destructive effect against cefotaxime and ceftriaxone than ceftazidime. CTX-M beta-lactamases are divided into five main groups based on amino acid changes, including CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 (10).
Therefore, considering the transfer of resistance genes by ESBL isolates and the identification of ESBL-producing isolates, it is very important to isolate these isolates to prevent treatment failure (14, 15). A simple and inexpensive phenotypic technique for isolating these isolates is the double-disc synergy test (DDST), which can be performed in many laboratories (14).
2. Objectives
The aim of the present study was to detect ESBLs-producing E.coli and K. pneumoniaee strains isolated from clinical specimens.
3. Methods
In this cross-sectional study, E. coli and K. pneumoniae strains were isolated in Shushtar hospitals, Khuzestan province (southwest Iran), from urine, blood, wound, and sputum specimens from March to October 2015. After culturing on Macconkey and Blood agar (Merck, Germany), colonies were identified by biochemical and microbiological test, including gram stain, oxidase, catalase, indol, motility, citrate, and TSI Urea, and lysine decarboxylase (16). The isolated bacteria were kept at -20°C in tryptic soy broth (TBS) plus 20% Glycerol.
3.1. Antimicrobial Sensitivity Pattern
The antibiotic sensitivity pattern was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines by disc diffusion test. In this method, ceftazidime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), aztreonam (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), amikacin (30 μg) (MAST, Germany) discs were used on Mueller- Hinton agar (MHA) (17).
3.2. ESBLs Phenotypic Screening
Double-disc synergy test (DDST) carrying out for isolation of ESBL-producing strains. In this test, ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime + clavulanic acid (30/10 μg), and cefotaxime + clavulanic acid (30/10 μg) were used on Mueller-Hinton agar. After incubation at 37°C for 18 hours, if the inhibition zone around the combination and single disc was ≥ 5 mm, the strain was considered ESBL positive. The positive and negative control isolates used in this test were K. pneumoniae ATCC 700603 and E. coli ATCC 25922, respectively (1).
3.3. Detection of bla TEM, bla SHV, and bla CTX-M Genes
Bacterial genomic DAN was extracted from all strains according to the manufacturer’s instruction (Sinaclon Co, Iran). Bla TEM, bla SHV, and bla CTX-M genes detected by polymerase chain reaction (PCR) assay using specific primers and DNA template (Table 1). Following the amplification, electrophoresis was carried out on 1.5% agarose gels for 50 min at 90 V that were visualized using a UV gel documentation system (BioRad, USA).
| Target Gene | Primer Sequence (5´- 3´) | Amplicon Size (bp) | Conditions | Volume Reactions | Reference |
|---|---|---|---|---|---|
| bla CTX-M | -TTTGCGATGTGCAGTACCAGTAA-; -CGATATCGTTGGTGGTGCCATA- | 590 | 1 cycle: 94°C; 4 min; 35 cycle: 94°C; 45 s; 56°C; 45 s; 72°C; 45 s; 1 cycle: 72°C; 7 min | 10X PCR Buffer: 2.5 μL, 10 mM dNTPs: 0.5 μL, 10 mM MgCl2:0.75 μL, 10 pmol F + R Primer: 1.25 μL, Taq DNA polymerase (5u μL): 0.2 μL, Template DNA: 5 μL; H2O up to 25 μL | (18) |
| bla TEM | -AGTGCTGCCATAACCATGAGTG-; -CTGACTCCCC GTCGTGTAGATA- | 431 | 1 cycle: 95°C; 5 min; 36 cycle: 95°C; 60 s; 54°C;.60 s; 72°C; 60 s; 1 cycle: 72°C; 5 min | 10X PCR Buffer: 2.5 μL, 10 mM dNTPs: 0.5 μl, 10 mM MgCl2: 0.75 μL, 10 pmol F + R Primer: 1.25 μL, Taq DNA polymerase (5u μL): 0.2 μL, Template DNA: 5 μL; H2O up to 25 μL | (19) |
| bla SHV | -GATGAACGCTTTCCCATGATG-; -CGCTGTTATCGCTCATGGTAA- | 214 | 1 cycle: 94°C; 5 min; 38 cycle: 94°C; 60 s; 53°C; 60 s; 72°C; 60 s; 1 cycle: 72°C;.7 min | 10X PCR Buffer: 2.5 μL, 10 mM dNTPs: 0.5 μL, 10 mM MgCl2: 0.75 μL, 10 pmol F + R Primer: 1.25 μL, Taq DNA polymerase (5u μL): 0.2 μL, Template DNA: 5 μL; H2O up to 25 μL | (19) |
3.4. Statistical Analysis
In this study, the statistical program (Statistical Package for the Social Sciences, SPSS) version 19 was used. Differences between proportions were analyzed using chi-square test, and P ≤ 0.05 was considered significant.
4. Results
In this study, one hundred E. coli and thirty K. pneumoniae isolates were isolated from different clinical samples. Table 1 shows the demographic characteristics of the strains isolated from clinical specimens. As shown in Table 2, the highest frequency of E. coli and K. pneumoniae was related to the intensive care unit (ICU) and emergency room, and most strains were isolated from urine samples.
| Variables | E. coli (N = 100) | K. pneumoniae (N = 30) |
|---|---|---|
| Gender | 76 (76) | 18 (60) |
| Female | ||
| Male | 24 (24) | 12 (40) |
| Clinical specimen | 84 (84) | 16 (53.47) |
| Urine | ||
| Blood | 12 (12) | 5 (16.6) |
| Sputum | 1 (1) | 5 (16.6) |
| Wound | 3 (3) | (13.3) |
| Sections | 29 (29) | 9 (30) |
| ICU | ||
| Emergency | 38 (38) | 9 (30) |
| Surgery | 8 (8) | 4 (13.3) |
| Internal | 25 (25) | 8 (26.7) |
a Values are expressed as No. (%).
The highest rates of antimicrobial resistance among E. coli isolates were obtained for cefotaxime 46 (46%), followed by ceftazidime 43 (43%), ceftriaxone 40 (40%), aztreonam 33 (33%), gentamicin 32 (32%), amikacin 25 (25%), and ciprofloxacin 23 (23%). On the other hand, the K. pneumoniae strains were resistant to ceftazidime 9 (30%), ceftriaxone 13 (43.5%), aztreonam 15 (50%), gentamicin 9 (30%), amikacin 13 (43.5%), and ciprofloxacin 7 (23.5%).
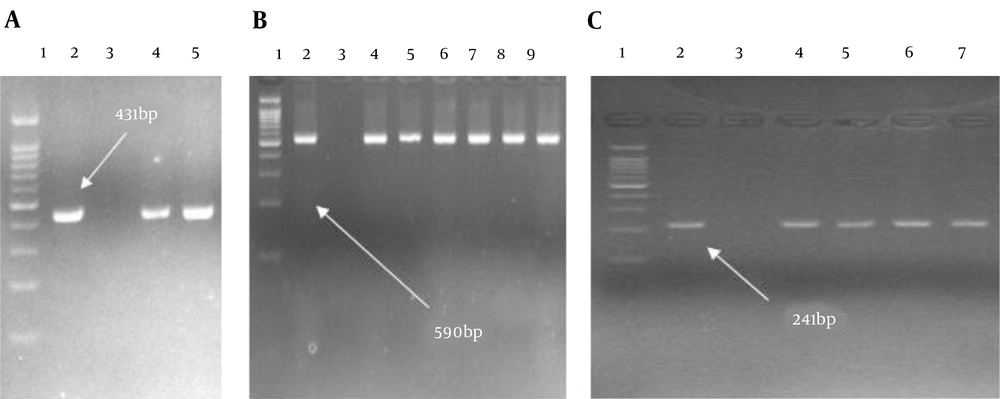
The overall frequency of ESBL-harboring K. pneumoniae and E. coli were 13.5% (4/30) and 28 (28/100) by DDST, respectively. According to the PCR assay results, among 28 ESBLs-producing E. coli, bla TEM (N = 28, 100%) was the most prevalent ESBL gene, followed by bla CTX-M(N = 26, 92.8%), and bla SHV (N = 1, 3.5%). Among four ESBL-producing K. pneumoniae isolates, 4 (100%), 4 (100%), and 3 (75%) carried bla CTX-M, bla TEM, and bla SHV genes, respectively (Figure 1).
Gel electrophoresis shows the PCR products of bla TEM, bla SHV, and bla CTX-M (A, Lane 1, DNA ladder 100 bp; Lane 2, control positive of bla TEM; Lane 3, control negative of bla TEM; Lanes 4 and 5, clinical isolates carrying bla TEM; B, Lane 1, DNA ladder 100 bp; Lane 2, control positive of bla CTX-M; Lane 3, control negative of bla CTX-M; Lanes 4 - 9, clinical isolates carrying bla CTX-M. C; Lane 1, DNA ladder 100 bp; Lane 2, control positive of bla SHV, Lane 3; control negative of bla SHV; Lanes 4 - 7, clinical isolates carrying bla SHV).
5. Discussion
K. pneumoniae and E. coli are the members of the Enterobacteriaceae family that cause nosocomial infections. Recently, in many parts of the world, antibiotic resistance due to the presence of beta-lactamase enzymes increased against a wide variety of antibiotics (20-22). In this study, the results indicate a high rate of antibiotic resistance, which is consistent with the results of many studies around the world, but in some studies, this rate of resistance is low, which indicates differences in the pattern of antibiotic use in different regions (23, 24). On the other hand, the frequency of ESBL strains was high in this investigation.
In Qureshi’s study et al. in Pakistan, 98.9, 96.7, and 93.4% resistance rates were described in E. coli against cefotaxime, ceftazidime, and cefuroxime (25). In Moayednia et al. study in Isfahan, E. coli clinical isolates showed 32, 38, and 33.4% resistance rate against ceftazidime, cefotaxime, and ciprofloxacin antibiotics (26). Given that the trend of antimicrobial resistance among clinical isolates in any geographical area is essential for recommendations in experimental antibiotic treatment of infections. The aim of this study was to determine the antimicrobial resistance and diagnosis of ESBL among E. coli and K. pneumoniae strains, which are dominant producers of ESBL. During the past decade, ESBL-producing E. coli and K. pneumonia isolates greatly differ worldwide. For example, at University Hospitals in Aleppo city, Syria, ESBL screened in E. coli and K. pneumoniae in abundance 62.89 and 67.5% (5). On the other hand, our result is higher compared to other reports 26.5% of E. coli and 43% of K. pneumoniae were ESBL-producing strains from Imam Reza Hospital of Mashhad, Iran (27). The frequency of ESBL-producing E. coli isolates was reported at 69.4% in Paris (28) and 41.6% in India (29).
In the current study, genotypic survey showed all isolates were positive for at least one of the studied genes on all phenotypically ESBL-producing E. coli and K. pneumoniae strains. Bla TEM was revealed in all tested isolates; so, bla TEM was the most prevalent gene in E. coli and K.pneumoniae ESBL-positive isolates. These results are in agreement with other studies, such as Wollheim et al., in southern Brazil bla TEM (95.3%) was the frequent gene followed by the bla CTX-M(82.8%), and the bla SHV gene (42.2%) among K. pneumoniae and 66.7, 50, 11.1% of ESBL-producing E. coli carrying bla TEM, bla CTX-M, and bla SHV gene, respectively (2). However, in AL-Subol study bla CTX-Mwas the commonest genotype (100% of K. pneumoniae and 76.14% of E. coli isolates), and followed by bla SHV was identified in K. pneumoniae (92.59%) and E. coli (57.95%) isolates (5). Peter Montso et al. in North-West Province South Africa detected bla TEM, bla SHV, and bla CTX-M genes in E. coli isolates (85.5, 69.6, and 58%, respectively) (30). Gautam study in India reported bla TEM gene was the most prevalent ESBL in E. coli and K. pneumoniae strains (31).
According to the results of this study, a high percentage of E. coli and K. pneumoniae isolated from Shushtar hospital patients produced ESBL and TEM genes. This indicates the need for changing the strategy of antibiotic therapy use to prevent transmission.
The difference in the prevalence of these genes in different studies may be due to differences in the pattern of use of antibiotics, including cephalosporins. Excessive use of antibiotics causes the spread of antibiotic resistance genes among bacteria, which can be prevented by changing the strategy of using antibiotics to transfer these genes among bacteria.
5.1. Conclusion
Because the bla TEM, bla SHV, and bla CTX-M genes are carried by plasmids, and these plasmids can carry other antibiotic resistance genes. They make it difficult to treat infections caused by antibiotic-resistant strains. Therefore, identification of ESBL-producing isolates can be effective in the treatment of these infections.