1. Background
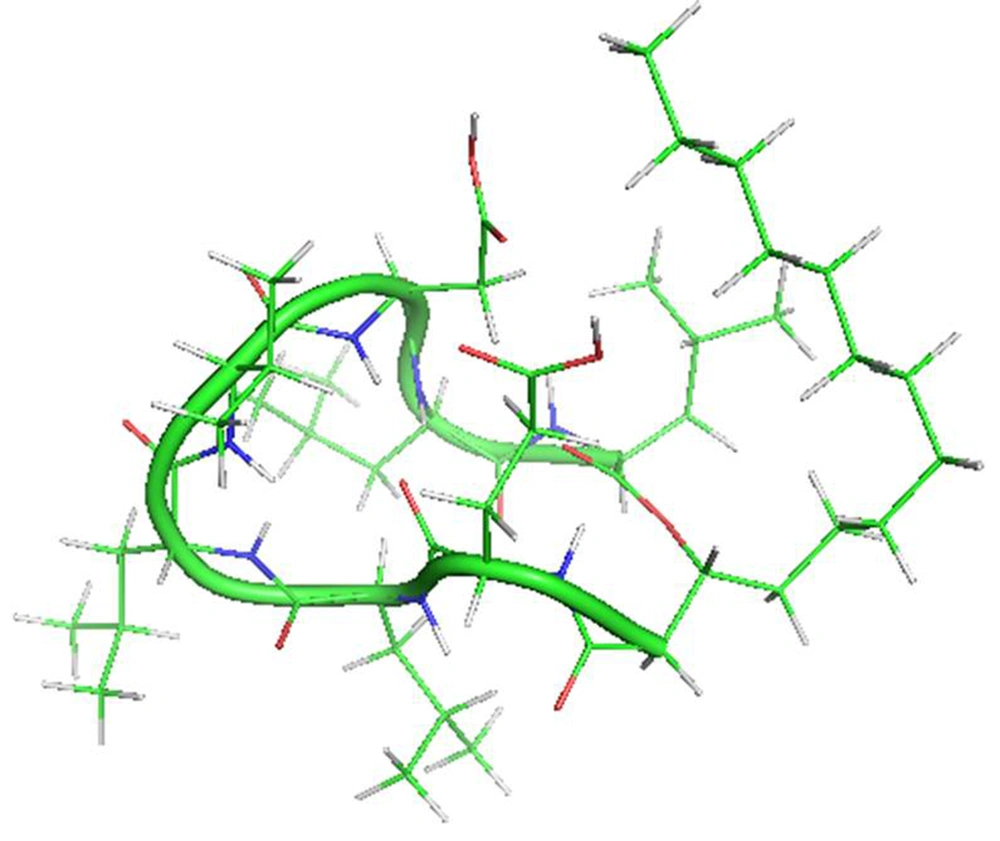
Lipopeptides are a particular group of cyclic peptides consisted of biomolecules with either a net negative (e.g., surfactin, daptomycin) or net positive charge (e.g., polymyxin). There are remarkable medicinal and biotechnological properties of these bioactive secondary metabolites (1-3). Surfactin is a cyclic amphipathic lipopeptide with a molecular weight of 1036 Da. A cyclic lactone ring arrangement with 12 to 16 carbon atoms shapes this heptapeptide (ELLVDLL) secondary metabolite, with the chiral series LLDLLDL interlinked with the chain lengths of β-hydroxy fatty acid (Figure 1). The residues of hydrophobic amino acids are found at situations 2, 3, 4, 6, and 7, though the residues of glutamyl and aspartyl at positions 1 and 5, respectively, give two negative charges to the molecule (4, 5). Surfactin adapts a beta-sheet structure with a distinctive horse-saddle conformation in an aqueous system and at the water/air interface, whose wide range of biological activities likely arises from this type of structure (6-8). During the stationary phase, surfactin is synthesized by various Bacillus species when nutrients are limited in culture media. By using different mutants lacking the synthesis of surfactin, a wide range of studies has been carried out on the role of this lipopeptide in bacterial physiology. In these strains, different functions are affected, such as swarming colonies in solid media (9, 10). Numerous potential functions have also been suggested: The enhancement of hydrophobic water-insoluble growth substrates in the surface area, resulting in higher bioavailability of nutrients and an effect on the binding and separation of bacterial cells to and from substrates (11, 12).
Besides its interfacial properties, surfactin exhibits several biological activities: (1) antibacterial and anti-inflammatory, (2) hemolytic, (3) antiviral, (4) anti-mycoplasma, (5) antitumoral, and (6) thrombolytic activities (13-16). Surfactin interacts with membranes and initiates lipid phase transitions as well as membrane destabilization (16). Various studies in the fields of genetics, biophysics, and biochemistry have revealed that phospholipids have numerous functions in different processes in cells. Their fundamental and prominent roles form a phospholipid bilayer as a permeability barrier to cells. This bilayer acts as a matrix and takes care of various proteins that have an essential function in cells, including energy or signal transduction, transport of solutes, DNA replication, targeting and trafficking of proteins, identification of cells, secretion, etc. (17). In every eukaryotic cell, the difference in head and aliphatic lipid chains leads to approximately 1,000 distinct lipid types (18, 19). Lipid compositions in prokaryotic and eukaryotic cell membranes are different from each other. Phosphatidic acid (PA), phosphatidylcholine (PtdCho), phosphatidylserine (PtdSer), phosphatidylethanolamine (PtdEtn), and phosphatidylinositol (PtdIns) are the most common structural lipids in eukaryotic cell membranes. Their tail part is diacylglycerol (DAG), which includes different lengths of saturated or cis-unsaturated fatty acyl chains. Among the mentioned phospholipids, in most eukaryotic membranes, PtdCho constitutes more than 50 percent of phospholipid membrane composition (18). However, regarding prokaryotic cells, the major structural lipid is phosphatidylglycerol (17, 20). Surfactin binding to different membranes and its mechanism of action can help us modify and optimize its structure in order to improve the efficacy of this lipopeptide in the future. For this purpose, we studied the interaction of this lipopeptide with two types of lipid bilayer models, including palmitoyl-oleoyl-phosphatidylglycerol (POPG) and 1-palmitoyl-oleoyl-glycero-phosphocholine (POPC) as eukaryotic and prokaryotic membrane models, respectively.
2. Methods
2.1. Preparation of Systems
The Surfactin lipopeptide coordinates file was downloaded from the protein data bank (PDB) (PDB ID: 2NPV). PRODRG server was used for generating the initial conformation of surfactin. Pre-equilibrated membranes were downloaded from the Tieleman laboratory and Lipidbook, including POPC (128 POPC lipids with 2460 water molecules) and POPG (128 POPG lipids with 128 Na+ and 3527 water molecules), respectively. To provide a sufficient aqueous phase for peptide positioning in the z-direction of membranes, approximately 5000 water molecules were added to the POPC and POPG systems. Both membrane systems were equilibrated in water for 100 ns before being used in molecular dynamics (MD) simulations. Using visual molecular dynamics (VMD) software (21), peptides were positioned in the water phase of the membrane. Water molecules were replaced by counter ions (Cl- and Na+) at the most positive/negative electrical potential in order to neutralize the system. Generally, four systems were designed.
2.2. MD Simulation Setup
The GROMACS software package, version 5.0.0, was used to perform all simulations (22-24). For the solvent and peptide, the GROMOSE96 force field parameters were used, and the updated GROMOS united-atom parameter was used after downloading the lipid.itp file from the Tieleman laboratory. Simulation systems were thermostatted at 310 K (25-27). In all three dimensions, periodic boundary conditions were used. Short-range and long-range electrostatic interactions were computed with a distance cutoff of 1.2 nm and the algorithm of particle mesh Ewald (PME), respectively (28). A cutoff of 1.2 nm was used to compute the van der Waals interaction. Energy minimization was applied to all systems using the steepest descent algorithm and tolerance of 500.0 kJ/mol/nm. By applying the Berendsen algorithm with a 0.1 ps coupling constant under the NVT-ensemble state condition for 200 ps, all simulation systems were equilibrated. In order to further equilibrate system components) peptide, lipid, and solvent (position restraints were placed on all heavy atoms of peptide using a 1000 kJ.mol-1.nm-2 spring constant, while the protein conformation was kept unchanged. Using the Grid-based searching method, the neighbor list was updated at a 10-step frequency, and the short-range adjacency list, a cutoff distance of 1.2 nm, was used. In all systems, the leap-frog algorithm was utilized with a two fs time step. Using the LINCS algorithm, all bonds were restricted (29).
Following the NVT equilibration, an isothermal-isobaric (NPT) equilibration was used for 1000 ps under standard pressure with a coupling constant of 0.5 ps applying the Parrinello-Rahman algorithm in semi-isotropic situations (Parrinello & Rahman, 1981). The temperature groups were coupled using the Nose–Hoover algorithm for NPT equilibration (Nos'e & Klein, 1983; Hoover, 1985). The same restrained position applied in the NVT step was also used during the NPT equilibration. All systems were submitted for unbiased MD runs after positional restraint equilibration. By starting from the same initial state, two independent 50-ns MD runs were performed using various initial velocity distributions for each system.
3. Results
3.1. Dynamics and Structural Changes of Surfactin
3.1.1. The Root-Mean-Square Deviation
Concerning the initial structure of the lipopeptide in the POPC and POPG membranes, the root-mean-square deviation (RMSD) is reported in Figure 2 as a function of time. For the surfactine-POPC simulation system, the lipopeptide RMSD was not stable during the simulation time, indicating that the system was not in an equilibrium structure. However, regarding the peptide part of surfactin in this simulation, the RMSD value increased to 0.29 nm at 6400 ps, and then reached equilibrated conditions up to the end of the simulation time. Also, the lipid part of molecules, similar to the whole lipopeptide structure, was not stable. So, we can say that the fatty acid chain of surfactin leads to its whole unstable structure in interaction with the POPC membrane. Regarding the surfactin-POPG simulation system, the whole lipopeptide structure, peptide part, and lipid part RMSDs stabilized at 0.38 nm, 0.33 nm, and 0.21 nm, respectively, in the first 5000 ps of the simulation time. Also, we can see that the RMSD of surfactin and its peptide part are very similar; therefore, the peptide part of the molecule leads to its whole stable structure in interaction with the POPG membrane.
3.1.2. Root-Mean-Square Fluctuations
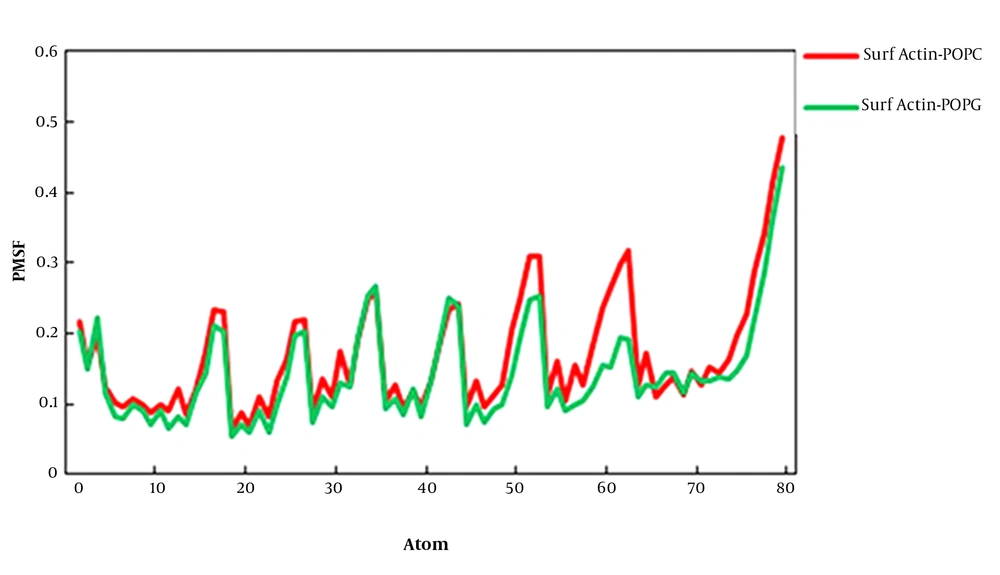
The root-mean-square fluctuations (RMSFs) of surfactin in the POPC and POPG membranes are studied as a function of atom number (Figure 3). The average atomic RMSFs for surfactin-POPC and surafctin-POPG were 0.167 nm and 0.142 nm, respectively. The average atomic RMSFs for peptide and the lipid part of surfactin in POPC were 0.132 nm and 0.104 nm, and in POPG were 0.114 nm and 0.097 nm, respectively. Thus, except for the terminal atoms of the fatty acid chain, the mobility of the lipid part was less than the peptide part, confirming the stability of the fatty acid chain in the membrane environment. In general, these plots indicated that the RMSF per atom for overall mobility of atoms for surfactin in two membranes were very similar, but in the POPG membrane, fluctuations of atoms were less than the POPC membrane.
3.2. Position and Orientation of Surfactin Into Membranes
3.2.1. Density
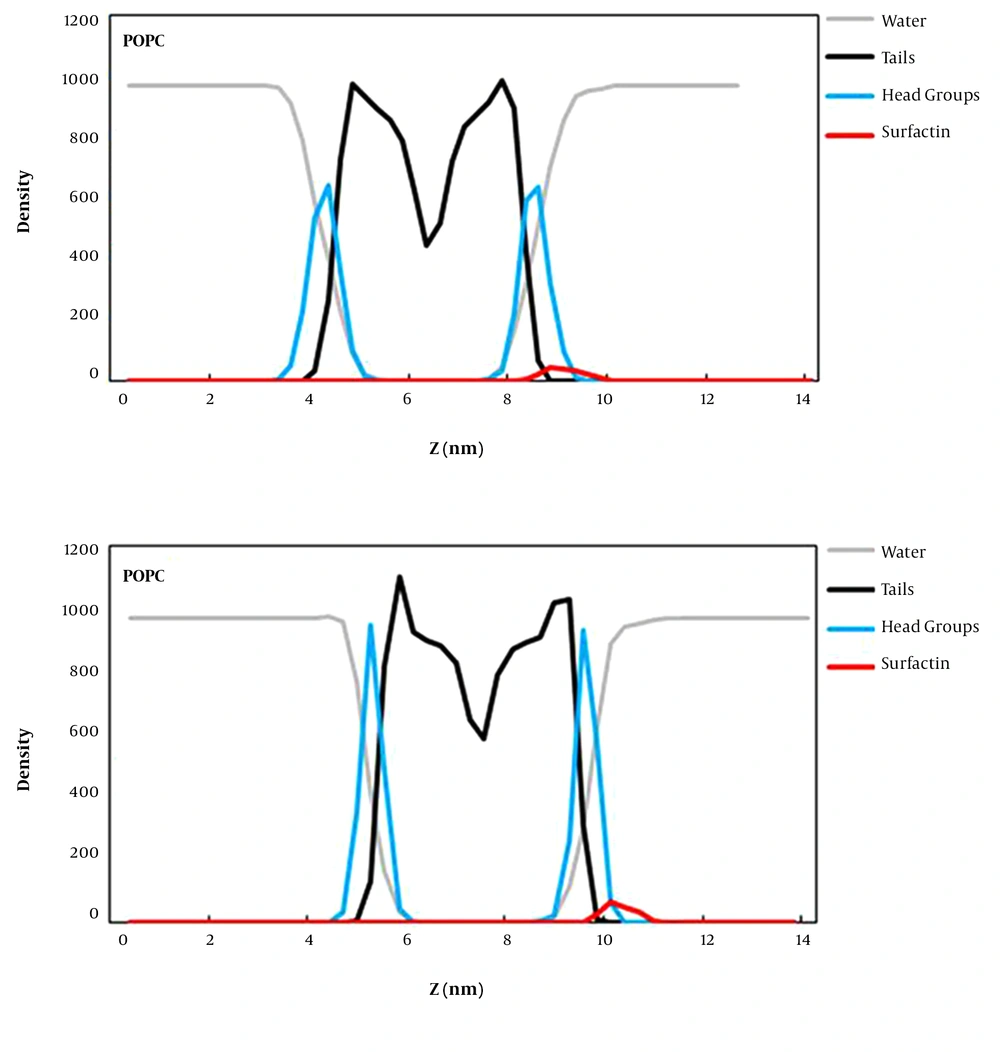
The density map along the z-axis of the simulation box for surfactin, water, head groups, and lipid tails was computed to determine the position of surfactin relative to the lipid surface and bulk solvent defining the water/bilayer interface (Figure 4). According to the obtained results, it can be understood that surfactin interacts with lipid head groups better than lipid tail regions of both membranes. These POPG bilayer interactions are more than the POPC bilayer’s, indicating that surfactin interacts properly with the anionic membrane (prokaryotic) compared to the zwitterionic bilayer. Also, some regions of this lipopeptide are still positioned within the aquatic phase of both simulation systems. So, it can be said that surfactin does not penetrate deeply into the membrane but interacts with the surface of membranes.
Hydrogen bonds (H-Bonds) one of the most essential factors in the mechanism of lipopeptides action is hydrogen bonds (H-bonds) and electrostatic interactions. Thus, we performed an H-bond analysis to elucidate interactions between surfactin and the lipid head groups of bilayer membranes. The average numbers of H-bonds between surfactin-water and surafctin-membrane in the first 10 ns for POPC were 31 and 0, and for POPG were 31 and 1, respectively. In the last 10 ns, these results were 27 and 1 for POPC, and 29 and 2 for POPG. In other words, the decreased rate of lipopeptide-water hydrogen bonds and the increased rate of lipopeptide-membrane hydrogen bonds were considered meager. Thus, in MD simulation time, surfactin formed slight hydrogen bonds with lipid head group that was not enough for penetrating membranes. These results are compatible with the density map, showing that the lipopeptide moves from the aquatic phase to the bilayer surface but does not penetrate membranes. Moreover, the tendency of surfactin for POPG (prokaryotic membrane) is more than POPC (eukaryotic membrane).
3.3. Influence of Surfactin on Membrane Structure
3.3.1. Order
The order parameter (-SCD) can be used for analyzing the influence of surfactin on the membrane structure. This analysis method compares the arrangement of lipid tails between the pure lipid bilayer and the peptide-associated bilayer (30, 31). This property of the membrane varies between different types. POPC and POPG are two types of the membrane with different charges and lengths of the head group region but the same hydrophobic tails. POPC is a zwitterionic lipid bilayer which is similar to a eukaryotic membrane environment, but POPG is an anionic lipid bilayer which is similar to a prokaryotic membrane environment. These lipids have an 18 carbon and a double-bond tail (oleoyl) and a 16 saturated carbon tail (palmitoyl) while POPC has a zwitterionic phosphocholine and POPG has an anionic phosphoglycerol as a head group (32).
The results of our study have indicated that the interaction of surfactin with the POPC membrane leads to increased (-SCD) values of sn1 and sn2 acyl chains rather than those of pure POPC simulation. In other words, the binding of surfactin to the POPC membrane causes the entire two fatty acid chains to become an order. Regarding the POPG membrane, the surfactin interaction significantly decreases the lipid acyl chain arrangement for a pure POPG bilayer. The presence of surfactin causes disordering of POPG acyl chains near to the lipid head groups more than the center of the bilayer, especially about sn1. Remarkably, this reduction in the arrangement of POPG fatty acid chains has shown that surfactin tends to destabilize the anionic membrane. Such data can provide valuable insights into antibiotic activities of surfactin.
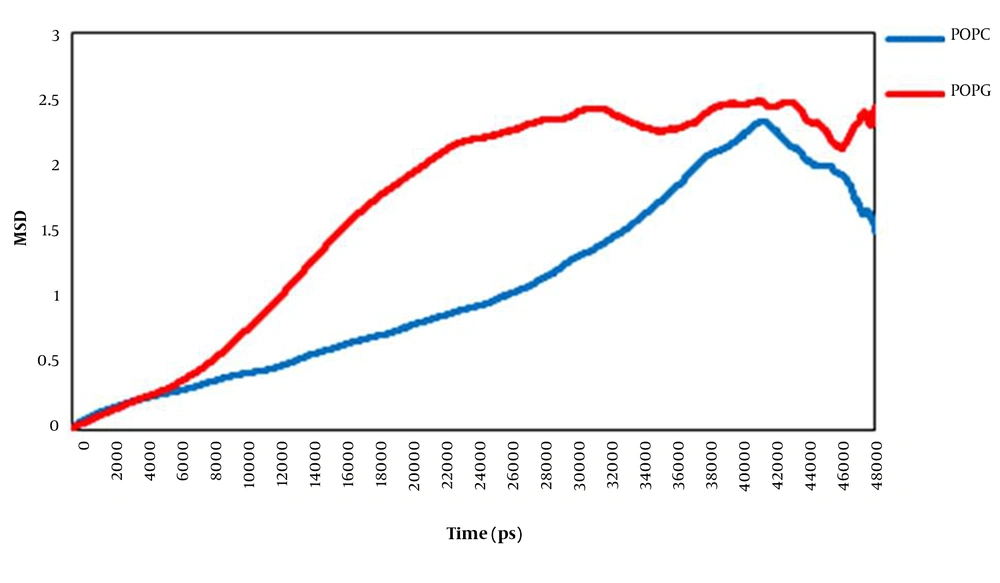
3.3.2. Mean Square Displacement
We have used mean square displacement (MSD) analysis to investigate the translational motion of lipids (Figure 5). The lateral diffusion rate of lipid molecules on the membrane surface is known to be a function of the membrane structure, the obstacle concentration, the temperature, the degree of hydration, and the time scale observed (33). The obtained data showed that the lateral displacement of the POPC molecules and the POPG lipid molecules were different. Plots indicted that in the presence of surfactin, the POPG membrane was more diffusive than the POPC bilayer. Consequently, it can be said that the movement of POPG molecules throughout the membrane is probably limited to the interactions between disordered POPG acyl chains.
4. Discussion
Surfactin is a cyclic heptapeptide that is closed by a β-hydroxy fatty acid chain. This lipopeptide has numerous attractive activities, including antibacterial, antiviral, antimycoplasma, hemolytic, and other different and powerful surface and interface activities (34). Several studies have demonstrated that surfactin's biological activity derives from the molecule's interaction with the membrane of target cells. Also, the composition of the phospholipid bilayer influences the action of this lipopeptide. Different suggestions such as membrane channel formation (35), detergent-like action (15), mobile carrier of cations (36) and modifiying action for the bulk physical properties of the membranes (37) have been reported as the mechanism of action for surfactin as a cyclic lipopeptide. Considering surfactin's diverse activities, it is essential to elucidate its cell selectivity and its mode of action at the atomic level.
In this work, we used MD simulations to study surfactin behavior in POPC and POPG bilayers as eukaryotic and prokaryotic membranes, respectively. Differences in how surfactin interacts with different types of membrane (membranes with different head groups and acyl chain lengths) were studied by experimental methods, including: (1) nuclear magnetic resonance (NMR); (2) dynamic light scattering (DLS); (3) differential scanning calorimetry (DSC).
X-radiation (X-ray) (38-41). Our obtained data regarding MD simulation as well as experimental studies showed that the tendency of surfactin for membrane depends on its composition. The tendency of this lipopeptide for the POPG bilayer is more than the POPC bilayer. Moreover, surfactin interacts peripherally with both membranes. This finding suggests that surfactin probably modifies membranes’ bulk physical properties. Also, the results of the order parameter revealed that surfactin does not have the same effects on the POPC and POPG membranes. Regarding POPC, the presence of surfactin caused more ordered acyl chains. However, regarding POPG, the acyl chains became disordered in the presence of this molecule. The results of the ordered analysis showed that the two fatty acid chains of POPC became ordered but regarding the POPG membrane, the two fatty acid chains became disordered. In addition, the results of MSD analysis showed that the movement of POPG molecules throughout the membrane was limited. This is probably because of the interactions between disordered POPG acyl chains. All these data indicated that the interaction of surfactin with the membrane led to the destabilized membrane, especially the POPG membrane. Consequently, these two types of lipid bilayers have different responses to surfactin binding. These results show that surfactin preferentially interacts with prokaryotic membranes to modify its molecular order.