1. Background
Systemic lupus erythematosus (SLE) is one of the complex diseases with a wide range of clinical criteria, including antinuclear antibody (ANA) and anti-double-stranded DNA (anti-dsDNA) antibodies (1-3). The incidence of SLE disease varies between different populations. The cause of SLE disease is unknown. Environmental and genetic factors are among the risk factors for SLE. The prevalence of SLE is high in Asians and notably in Iranian populations (4, 5). Recently, many efforts have been made to find a link between SLE susceptibility and genetic variants (6-9). The tumor necrosis factor α (TNF-α) gene produces an inducible pro-inflammatory cytokine (10, 11), which is fixed in human chromosome 6 within the major histocompatibility complex (MHC) class III region (12). It seems that the TNF-α gene is connected to the pathogenesis of inflammatory disorders (13, 14). Several studies on different diseases have shown the effect of single-nucleotide polymorphisms (SNPs) on the TNF-α promoter region (15, 16). However, studies have not definitely determined the association between TNF-α promoter gene SNPs and SLE (17-20). Rs18008629 at position -308 G/A polymorphism is related to increased potential and intensity in a variety of autoimmune diseases (19, 21-23). The second polymorphism, a common functional polymorphism is located at position -863 C/A (24). Several studies have analyzed the association between rs1800630 at the position -863 C/A TNF-α promoter gene polymorphism and inflammatory disorders such as SLE (18, 25-28). However, different populations have shown different results. No studies have been performed to study the association with rs18008629 at position -308 G/A and also rs1800630 at position -863 C/A TNF-α gene polymorphisms in the case of SLEin the Iranian Lor Population.
2. Objectives
We studied the association between rs1800629 TNF-α at position -308 G/A and rs1800630 TNF-α at position -863 C/A promoter polymorphisms and the susceptibility of SLE hazard.
3. Methods
3.1. Patients
According to the American College of Rheumatology (ACR) criteria, 120 unrelated SLE patients were selected. Also, 120 healthy individuals with no personal and family history of autoimmune disease were selected as controls. Both serological factors (ANA autoantibodies and anti-dsDNA) associated with the disease were also determined by diagnostic tests in patients, such as ANA autoantibodies (n = 114) and anti-dsDNA (n = 116). For this study, we selected all cases and controls from the Iranian Lor population.
3.2. Genotyping
Genomic DNA was isolated from peripheral blood leukocytes using the salting-out method (29). Briefly, 500 µL of blood was transferred to 1.5-µL microfuge tubes, and 1-mL cold water was added. The solutions were gently mixed and centrifuged at 13 000g for 1 minute at room temperature. Then, the supernatant was discarded. The procedure was repeated twice. Next, 300 µL TES buffer (pH = 7.5; NaCl [150mM], Tris-base [10mM], EDTA [10mM]), 320 µL SDS 10%, and 25 µL proteinase K (CinnaGen, Iran) were added, and the mixture was incubated at 37°C for 2 hours. Then, 220 µL of saturated NaCl was added with gentle mixing, and the mixture was centrifuged at 13 000g for 15 minutes. The supernatant was transferred to a new microfuge tube, where 550 µL of cold isopropanol was added and centrifuged at 13000 g for 2 minutes. The supernatant was discarded, and 1 mL of cold ethanol 70% was added. The suspension was gently mixed and centrifuged at 13000 g for 1 minute. Finally, pellets were dried before dissolving in 50 µL of TE buffer (Tris base [10mM], EDTA [1mM]) and preserved at -20°C. Recognition of TNF-α -308 and -863 polymorphisms was performed by the tetra-primer amplification-refractory mutation system (tetra-primer ARMS)–polymerase chain reaction (PCR) method. Four primers were tested, of which 2 inner primers (ie, inner forward and reverse) and 2 outer primers (ie, the outer forward and reverse) were the same primers. All 4 primers were designed using the NCBI bioinformatics database and then blasted (Table 1).
| Name | Primer Sequence | Tm | PCR Product |
|---|---|---|---|
| Rs1800629 | |||
| Outer forward | 5′- GGACCCAAACACAGGCCTCAG -3′ | 60.2 | 323 bp |
| Outer reverse | 5′- TCCTCCCTGCTCCGATTCC-3′ | 61.2 | |
| Inner forward | 5′- GGCAATAGGTTTTGAGGGCGAGGG-3′ | 62.2 | 217 bp |
| Inner reverse | 5′- GGAGGCTGAACCCCGTACT-3′ | 62.1 | 106 bp |
| Rs1800630 | |||
| Outer forward | 5′-GGCTCTGAGGAATGGGTTAC-3′ | 57.67 | 200 bp |
| Outer reverse | 5′-TGGCCATATCTTCTTAAACGT-3′ | 55.01 | |
| Inner forward | 5′-TCGAGTATGGGGACCCCCA-3′ | 60.46 | 121 bp |
| Inner reverse | 5′- ATGGCCCTGTCTTCGTTAAGG-3′ | 62.5 | 158 bp |
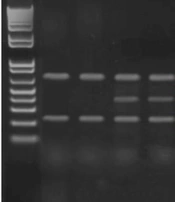
As a positive control, 323-base pairs (bp) and 200-bp constant DNA fragments (for TNF-α -308 and -863 polymorphisms, respectively) were amplified with outer primers. For the TNF-α -308 polymorphism, the thermal cycle of the test was as follows 94°C(4 minuts) (primary denaturation), annealing (for 30 cycles): 94°C (denaturation), 56°C(annealing), and 72°C (30 s each) (extension) and final elongation 72°C (7 minuts). For the TNF-α -863 polymorphism, the thermal cycling condition was followed by primary denaturation at 94°C (4 minutes), annealing (for 30 cycles): 95°C (denaturation), 60°C (annealing), and 72°C (30 s each) (extension) and final elongation 72°C (7minuts). Then, the products were visualized using electrophoresis in 2% and 2.5% agarose gels (TNF-α -308 and -863 polymorphisms, respectively) and stained with DNA safe stain (CinnaGen, Iran; Figure 1).
Tetra-primer amplification-refractory mutation system–polymerase chain reaction analysis of TNF-α (-308 G/A [A] and -863 C/A [B]) gene polymorphisms: Lane 1-A: 50-bp DNA ladder. Lanes 2 and 3: GG genotype. Lanes 4 and 5: GA genotype. Lane 3-B: 50-bp DNA ladder. Lanes 2 and 5: AA genotype. Lanes 1 and 4: CA genotype.
3.3. Statistical Analysis
The distribution of rs1800629 and rs1800630 genotypes was checked to analyze the deviation from Hardy-Weinberg equilibrium in SLE cases and controls using the chi-square test. Using chi-square and logistic regression tests, allelic and genotypic dispensation between the patients and healthy controls was analyzed. P-values less than 0.05 were considered statistically significant. The odds ratio (OR) and 95% CIs were also evaluated. Besides these 2 polymorphisms, the eventual correlation with 2 clinical manifestations was examined by the chi-square test. Statistical analysis of the data was performed using SPSS version 24 (SPSS Inc, Chicago, Ill, USA).
4. Results
In this study, TNF-α -308 G/A and -863 C/A SNPs were analyzed in 120 healthy controls and 120 cases with SLE. The -308 G/A polymorphism showed a significant departure from Hardy-Weinberg equilibrium among controls and patients in genotype distribution (P < 0.0001). Genotype distribution analysis for -863 C/A polymorphism Hardy-Weinberg equilibrium showed a significant deviation among patients but not controls (P < 0.0001 and P > 0.05, respectively).
4.1. Characteristics of Cases and Controls
The chi-square test (χ2 test) showed no significant association between gender and disease incidence (P = 0.244). The t-test for both independent samples indicated that age dissimilarity observed between the patient and control groups was statistically significant (P = 0.01). It means that the average age was higher in the control group than in the patient group. However, since this disease often occurs at an early age, it is concluded that our control subjects are perfectly matched for differentiation with our patients.
4.2. Allele and Genotype Frequencies of TNF-α Genetic Polymorphisms
For both TNF-α genetic polymorphisms, allele and genotype frequencies were calculated (Table 2). The -308 G/A SNP (rs1800629) allele frequency was not remarkably different. The frequency analysis of the A allele at position -863 of the TNF-α gene was remarkably higher in SLE cases than in healthy controls (OR = 3.426; 95% CI, 1.985 - 5.914). Accordingly, it is concluded that the AA genotype is correlated with an increased hazard of SLE disease (OR = 4.489; 95% CI, 2.464 - 8.177; P < 0.0001).
| Gene Name SNP Database ID (Cucleotide Change) | SLE | Controls | P-Value | Odds Ratio (95% CI) |
|---|---|---|---|---|
| rs1800629 (-308G>A) | 0.341 | 1.322 (0.744 - 2.347) | ||
| A/A | 30 (0.25) | 35 (31.7) | ||
| G/A | 90 (0.75) | 85 (68.3) | ||
| G/G | 0 | 0 | ||
| Alleles | 1.178 (0.810 - 1.712) | |||
| A | 150 (62.5) | 158 (65.83) | 0.391 | |
| G | 90 (37.5) | 82 (34.167) | ||
| rs1800630 (-863C>A) | < 0.0001 | 4.489 (2.464 - 8.177) | ||
| A/A | 63 (52.5) | 101(84.17) | ||
| C/A | 57 (47.5) | 19 (15.84) | ||
| C/C | 0 | 0 | ||
| Alleles | 3.426 (1.985 - 5.914) | |||
| A | 183 (91.67) | 220 (76.25) | < 0.0001 | |
| C | 57 (7.916) | 19 (23.75) |
Abbreviations: TNF-α, tumor necrosis factor α; SLE, systemic lupus erythematosus.
a Values are expressed as No. (%). P-values less than 0.05 were considered statistically significant.
4.3. TNF-α Genetic Polymorphisms and Clinical Features of SLE
The possible association between rs1800629 and rs1800630 TNF-α SNPs and both clinical features of SLE patients (ie, ANA [n = 114] and anti-dsDNA antibodies [n = 116]) were analyzed (Table 3). No statistically significant correlation was observed between these 2 polymorphisms and the clinical features of SLE patients.
| Serological Features | ||||||
|---|---|---|---|---|---|---|
| ANA (+) | ANA (-) | P-Value | Anti-dsDNA (+) | Anti-dsDNA (-) | P-Value | |
| Rs1800629 | ||||||
| Alleles | 0.390 | 0.543 | ||||
| A (%) | 100 (43.5) | 104 (45.61) | 99 (42.67) | 109 (46.98) | ||
| G (%) | 14 (6.141) | 10 (4.39) | 13 (5.61) | 11 (4.75) | ||
| Genotypes | 0.653 | 0.517 | ||||
| AA (%) | 43 (37.72) | 47 (41.22) | 43 (37.06) | 49 (42.25) | ||
| GA (%) | 14 (24.57) | 10 (8.78) | 13 (11.20) | 11 (9.48) | ||
| Rs1800630 | ||||||
| Alleles | 0.875 | 0.658 | ||||
| A (%) | 87 (38.16) | 88 (38.59) | 85 (36.64) | 94 (40.52) | ||
| C (%) | 27 (11.48) | 26 (11.41) | 27 (11.64) | 26 (11.20) | ||
| Genotypes | 0.851 | 0.589 | ||||
| AA (%) | 30 (26.32) | 31 (27.19) | 29 (25) | 34 (29.31) | ||
| CA (%) | 27 (23.68) | 26 (22.8) | 27 (23.28) | 26 (22.42) | ||
5. Discussion
In different ethnic groups, studies have suggested a correlation between TNF-α polymorphisms and SLE risk (30, 31). However, the effect of the TNF-α polymorphism on the ability of SLE disease is yet unclear. Some studies have shown that the TNF-α -308 A allele has a significant transcriptional effect, but others have claimed that this polymorphism has no effects on TNF-α function (31-35). The results of this study confirmed the association between the TNF-α -308 G/A allele and SLE, as it has been found in most populations, including Taiwanese patients (25). The current study indicated that none of the genotype and allele frequencies of the re1800629 polymorphism at position -308 G/A were remarkably associated with Lor SLE patients compared to controls. Generally, the results of association studies on different populations between re1800629 and rs1800630 and susceptibility of SLE risk are different. In Caucasian SLE patients, Tsuchiya et al showed that -863A, -308G haplotypes were associated with disease intensity (36), whereas McHugh et al showed no significant association between -863A, -308G and disease risk (37). Our results suggested that allele and genotype frequencies of rs1800630 were significantly associated with SLE in the Iranian Lor population. Functional analysis of the rs1800630 polymorphism in the promoter domain of TNF-α at position -863 showed contradictory results. Although the association between TNF-α gene polymorphisms and SLE is ambiguous in different ethnic histories, the -863 C allele may play a role in susceptibility to SLE in the Lor population, partially through their higher promoter occupation of TNF-α production.
Our statistical analysis showed no significant relationship between anti-dsDNA and ANA with the type of the genotype. Consequently, further studies are required in this respect between various Iranian populations and the increased potential of SLE hazard.
5.1. Conclusions
This study showed that TNF-α -863 SNP was associated with SLE in the examined patients. No significant association was observed between clinical features of SLE patients and these TNF-α promoter gene polymorphisms. This indicates that further studies with larger sample sizes on different populations are needed to find the exact role of this gene.
![Tetra-primer amplification-refractory mutation system–polymerase chain reaction analysis of <i>TNF-α</i> (-308 G/A [A] and -863 C/A [B]) gene polymorphisms: Lane 1-A: 50-bp DNA ladder. Lanes 2 and 3: GG genotype. Lanes 4 and 5: GA genotype. Lane 3-B: 50-bp DNA ladder. Lanes 2 and 5: AA genotype. Lanes 1 and 4: CA genotype. Tetra-primer amplification-refractory mutation system–polymerase chain reaction analysis of <i>TNF-α</i> (-308 G/A [A] and -863 C/A [B]) gene polymorphisms: Lane 1-A: 50-bp DNA ladder. Lanes 2 and 3: GG genotype. Lanes 4 and 5: GA genotype. Lane 3-B: 50-bp DNA ladder. Lanes 2 and 5: AA genotype. Lanes 1 and 4: CA genotype.](https://services.brieflands.com/cdn/serve/3170b/ae9b12441125ecab55f1308c0844dd187c1654cf/gct-115542-g001-F1-preview.webp)