1. Background
Methamphetamine addiction is a chronic debilitating cerebral disorder and is considered one of the costliest disorders worldwide. In the last decade, it has been reported that approximately 34,000,000 individuals have used methamphetamine in their life in the world (1). The methamphetamine was easily synthesized by relatively inexpensive compounds and was a highly addictive psychostimulant (2).
So far, limited studies have been performed on molecular pathogenesis of methamphetamine addiction, and efficient diagnostic and therapeutic methods have not been identified. In recent years, methamphetamine abuse disorder is commonly diagnosed using liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) methods through identification of methamphetamine metabolites from whole blood, plasma, and serum samples (3). However, the mentioned methods are expensive and time-consuming, which require various equipment and materials. Therefore, development of an inexpensive and reliable approach for rapid diagnosis and efficient treatment approach for methamphetamine abuse disorder is still of great importance (4).
Several small non-coding oligonucleotides, microRNAs (miRNAs) are identified, which play important roles in the regulation of gene expression during brain differentiation and development (5, 6). Recently, a lot of diversity of miRNAs have been demonstrated that are involved in mature neurons as well as development and differentiation of neurons. Previous studies reported that miRNAs play an important role in morphogenesis, memory formation as well as drug addiction (7, 8). Evidence suggests that miRNAs are involved in the pathogenesis of various drugs abuse, such as cocaine, heroin, alcohol, and nicotine (9, 10).
Evidence also suggests that methamphetamine is one of the most commonly used illegal drugs in the world. In addition, the number of people dye from methamphetamine use has significantly increased in recent years (1, 2). Therefore, it is important to identify the underlying molecular mechanisms of methamphetamine abuse disorder to find specific biomarkers for therapeutic approaches.
2. Objectives
Both miR-186 and miR-195 play an important role in the regulation of several molecular targets in the human brain such as angiogenesis, inflammation, synaptic transmission, and neurological mediators development (11, 12). However, molecular mechanisms of miRNA-186 and miRNA-195 function are unclear in Iranian patients with methamphetamine addiction. Here, we investigated the expression levels of miRNA-186 and miRNA-195 in the whole blood of Iranian patients with methamphetamine abuse disorder.
3. Methods
3.1. Study Subjects
In the present case-control study, we enrolled 60 patients with methamphetamine abuse disorder, newly diagnosed without any treatment, from addiction treatment centers in Tabriz, Iran from 2018 to 2019. Moreover, 60 healthy individuals without any drug addiction were enrolled as the control group. The individuals with major psychiatric disorders, chronic disorders, cardiovascular disease, and brain disease were excluded from this study. All subjects, age- and ethnically-matched, were enrolled from population of East Azerbaijan province, Iran. Demographic information, clinical characteristics, and lifestyle (age, gender, marital status, literacy levels, drug use history, and syphilis infection status) of all subjects were recorded through interviews. All members of case and control groups signed a consent form according to Declaration of Helsinki ethical standards, approved by the Ethics Committee of Islamic Azad university, Tabriz Branch (IR.IAU.TABRIZ.REC.1398.082).
3.2. Quantitative Real-time PCR
The whole blood samples (5 mL) were obtained from all members of case and control groups after 12 hours of fasting. Extraction of total RNA from whole blood samples was conducted using RNA extraction kit (GeneAll Biotechnology, Germany), according to instruction of the manufacturer. Poly-A polymerase enzyme was used to polyadenylation of the extracted RNA samples under the following conditions: 37°C for 30 minutes and then 65°C for 20 minutes. Next, the BON-RT adaptor primers were used to synthesize cDNA under the following conditions: 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes, and was stored at 4°C. The expression of miRNA-186 and miRNA-195 was investigated in both case and control groups in triplicate using a TaqMan probe-based RT-qPCR assay, in 15 μL total volume: PCR buffer (7.5 μL), cDNA (1.5 μL), forward primer (0.5 μL), reverse primer (0.5 μL), and DEPC-treated RNase free water (4.5 μL), under the following conditions: 94°C for 1 minute (initial denaturation), 94°C for 10 seconds (denaturation), 94°C for 30 seconds (annealing), and 72°C for 20 seconds (extension). The used primers were Has-miR-186-F-5'-CGGCAAACAATTCTCC-3' and Has-miR-195-F-5'-TTGGTAGCAGCACAGAAA-3'. The threshold cycle (CT) was identified, and the expression of miRNA-186 and miRNA-195 was normalized to U6 as endogenous control and calculated using the 2−ΔCq method.
3.3. Statistical Analysis
Statistical analysis was performed with SPSS software (version 21.0) and GraphPad Prism 6 software. The results of demographic information, clinical characteristics, lifestyle, and miRNA expression are presented as mean ± standard error of mean (SEM) or mean ± standard deviation (SD). Difference in the expression of miRNAs in the case and control groups was evaluated using Pearson’s correlation analysis. In addition, difference in demographic information, clinical characteristics, lifestyle of the case and control groups were evaluated using Chi-square and independent sample t-test. A P-value < 0.05 was considered statistically significant.
4. Results
4.1. Participants’ Characteristics
The statistical analysis demonstrated a significant difference between patients with methamphetamine addiction and healthy controls in terms of age, marital status, and syphilis infection (P < 0.05). However, the difference between patients with methamphetamine addiction and healthy controls was not significant in terms of educational degree and BMI (P > 0.05). The demographic information, clinical characteristics, and lifestyle of the studied patients with methamphetamine addiction and healthy controls are summarized in Table 1.
| Variable | Case (N = 60) | Controls (N = 60) | P-Value b |
|---|---|---|---|
| Age (y) | 28.41 ± 2.51 | 32.73 ± 9.22 | < 0.001 |
| BMI (kg/m2) | 22.19 ± 2.18 | 22.34 ± 2.55 | 0.529 |
| Marital status | 0.008 | ||
| Married | 28 (46.6) | 42 (70.0) | |
| Single | 18 (30.0) | 12 (20.2) | |
| Divorced | 14 (23.3) | 6 (10.0) | |
| Educational degree | 0.265 | ||
| Under diploma and diploma | 44 (73.3) | 36 (60.0) | |
| Higher diploma | 16 (26.6) | 24 (40.0) | |
| Drug use history | |||
| Onset age of drug use (years) | 24.78 ± 2.28 | - | - |
| Drug use time (y) | 4.56 ± 3.24 | - | - |
| Times of addicts drug per day | 1.87 ± 2.11 | - | - |
| Drug manner | |||
| Injection | 4 (6.6) | - | - |
| Oral inhalation | 56 (93.3) | - | - |
| Syphilis infection status | < 0.001 | ||
| Positive | 8 (13.3) | 0 (0.0) | |
| Negative | 52 (86.6) | 60 (100.0) |
Abbreviations: BMI, body mass index.
a Data are presented as mean ± SD and No. (%).
b The P < 0.05 is statistically significant.
4.2. Expression of miRNAs
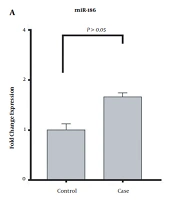
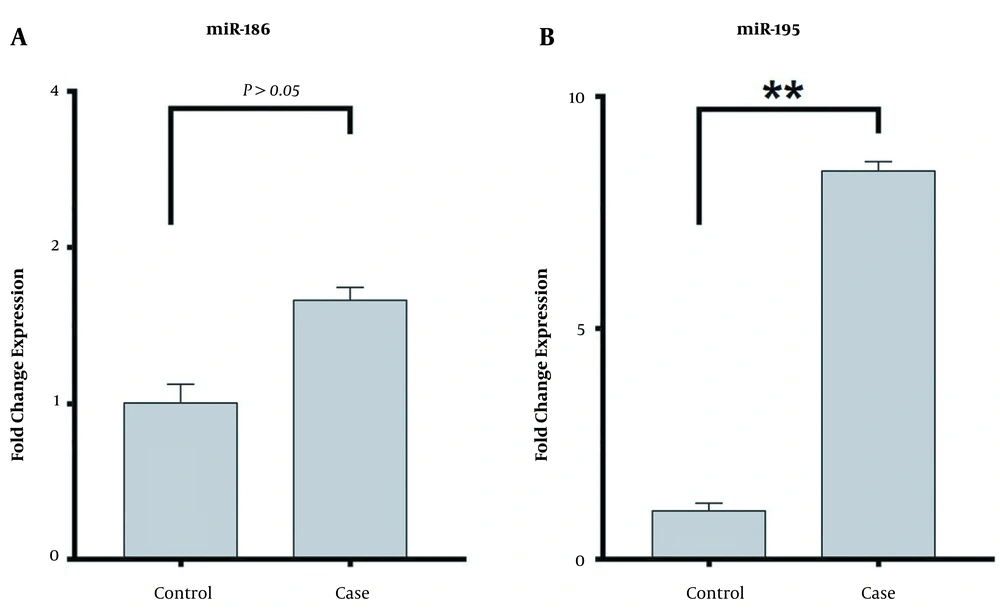
The present study demonstrated that the expression level of miRNA-195 significantly increased (8.75-fold change) in patients with methamphetamine addiction compared to healthy controls (P < 0.05). However, the expression level of miRNA-186 increased insignificantly (1.61-fold change) in the case group compared to the healthy controls (P > 0.05). The expression of miRNA-186 and miRNA-195 in the patients with methamphetamine addiction and healthy controls are shown in Figure 1.
4.3. Diagnostic Potential of miRNAs
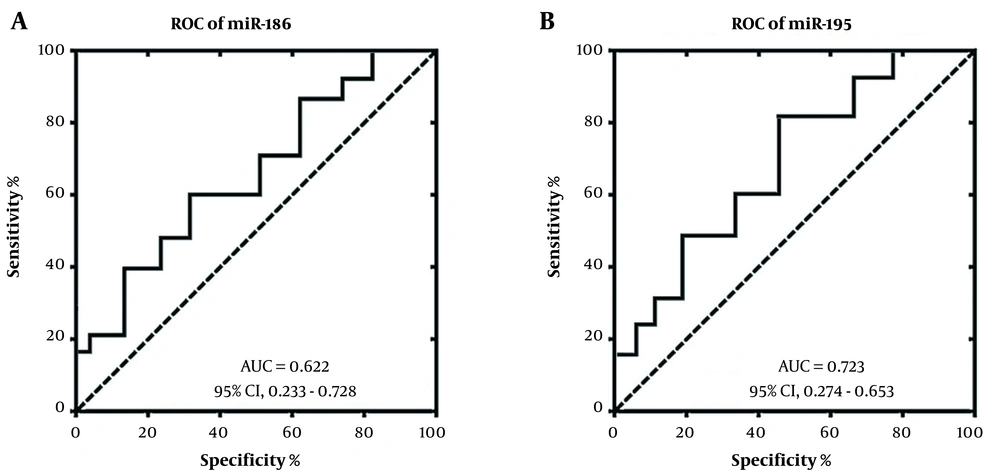
The receiver operating characteristics (ROC) curve analysis was used to investigate diagnostic potential of miRNA-186 and miRNA-195 for patients with methamphetamine addiction. The ROC curve analysis demonstrated that the area under the curve (AUC) score for miRNA-186 was 0.622 to discriminate patients with methamphetamine addiction from healthy controls (Figure 2A). In addition, the AUC score was 0.723 for miRNA-195 to discriminate patients with methamphetamine addiction from healthy control (Figure 2B). Our study showed that the miRNA-195 could be used as a reliable biomarker for diagnosis or prognosis of patients with methamphetamine addiction.
5. Discussion
Evidence suggested that dysregulation of miRNAs played important roles in neuronal function, neurogenesis, as well as neurobiological processes (synaptic plasticity) (13-15). In addition, miRNAs are involved in addiction progress through direct modification of synaptic remodeling, dendritic spine morphogenesis, rewarding properties of drugs, self-administration rates of alcohol, and drug-seeking behavior (14, 16). Previous studies reported that miRNAs in blood and brain could be used as an accurate and reliable biomarker for diagnosis of addiction (17, 18). Currently, several miRNAs have been considered biomarkers for diagnosis and prognosis of several mental and physical disorders (19, 20). However, the expression level of various miRNAs in subjects with methamphetamine addiction is largely unclear. Therefore, identification of addiction-related miRNAs can be used for development of novel diagnosis and therapeutic approaches in patients with methamphetamine addiction.
In this study, we investigated the expression of miRNA-186 and miRNA-195 in whole blood samples of patients with methamphetamine addiction. We showed that the expression of miRNA-195 significantly increased (8.75-fold change) in patients with methamphetamine addiction compared with healthy controls, whereas the expression level of miRNA-186 insignificantly increased in patients with methamphetamine addiction. Moreover, we demonstrated that altered expression of miRNA-195 provides an accurate and reliable biomarker for rapid diagnosis of methamphetamine addiction in forensic and clinical applications.
Previous studies suggested that the expression level of miRNAs in whole blood samples is associated with methamphetamine and other drugs addiction (20-22). In a most recent study by Gu et al., it was demonstrated that the serum levels of miRNA-9-3p significantly increased in patients with methamphetamine addiction (21). In another study by Zhao et al., the plasma levels of miRNA-181a, miRNA-15b, miRNA-let-7e, and miRNA-let-7d, significantly decreased in patients with methamphetamine addiction (22). In a study by Zhang et al., the blood levels of miRNA-181a significantly decreased in patients with methamphetamine addiction compared to healthy controls (23). We also showed significant alteration in the expression of the miRNA-195 in blood samples of patients with methamphetamine addiction compared to healthy controls. The mentioned studies may provide the important role of miRNAs in regulating methamphetamine abuse disorder. However, molecular mechanisms, as well as physiological and pathological role of miRNAs in patients with methamphetamine addiction, remain unknown. Previous studies suggested that addiction-related miRNAs played an important role in neurological and psychological disorders through involving various signaling pathways, including CREB, MAPK, G-Protein, GnRH, and Couple Receptor (23, 24). In this regard, identification of exact roles of miRNA-186 and miRNA-195 in pathological and physiological process in patients with methamphetamine addiction requires further investigation. In addition, further studies are required to identify the miRNAs as biomarkers for diagnosis and prognosis of patients with methamphetamine addiction.
5.1. Conclusions
Generally, we suggest that miR-195 may play an important role in pathology of methamphetamine addiction and can be considered an appropriate blood biomarker for diagnosis of patients with methamphetamine addiction. However, further studies are required to identify the exact role of miR-186 and miR-195 in neurological signaling pathways in patients with methamphetamine abuse disorder.