1. Background
Acinetobacter baumannii is a gram-negative, aerobic, nonfermenting, and rod-shaped ubiquitous in the medical environment and is generally regarded as a significant opportunistic pathogen (1). One of the public health threats that have recently been considered in the United States, Europe, Asia, and the Middle East is the rapid increase in the antibiotic-resistant isolates of A. baumannii (2). This pathogen is responsible for many infections, such as bacteremia, urinary tract infection (UTI), and respiratory tract infections, especially in immunocompromised patients (3). Acinetobacter baumannii transmits and survives in the hospital by attaching to different surfaces, such as cerebrospinal fluid shunts and vascular catheters. Catheter-acquired UTIs (CAUTIs) are among the most common nosocomial infections. In the previous study, forming A. baumannii biofilms along the catheter surface was the most important cause of bacteriuria (4).
Acinetobacter baumannii has several virulence factors, among which the ability to form biofilm is one of the most important factors (5). Biofilms are complex bacterial communities attached to surfaces, created by an extracellular matrix produced by bacteria. This matrix comprises polysaccharides, DNA, and proteins (5). Biofilm formation is a complex process that requires many factors, including aggregation, collagen adhesion, pili expression, and iron uptake (6, 7). Among the diverse factors effective in biofilm formation, the biofilm-associated protein (Bap, high-molecular-weight proteins) encoded by the bap gene has a significant role in attachment to bronchial cells, structural integrity, and water channel formation in the biofilm (8). This protein is located on the outer surface of bacteria and consists of a central core of the successive iterations of similar sequences (9). Disruption of the bap gene reduces the thickness and volume of biofilm and interbacterial cell adhesion (10).
2. Objectives
As mentioned, the bap gene is essential in forming A. baumannii biofilm. Therefore, our investigation aimed to assess the expression of the bap gene in A. baumannii using real-time polymerase chain reaction (PCR) in clinical samples from Khorramabad, Iran.
3. Methods
3.1. Sample Collection
This cross-sectional study was conducted during April 2017 - April 2018 on 43 A. baumannii strains from clinical samples, including urine, blood, wound, tissues, and chest sputum collected from teaching hospitals in Khorramabad (west of Iran). All strains were identified by microbiological and biochemical tests, such as oxidase, oxidation-fermentation (OF), triple sugar iron (TSI) test, motility in sulfur, indole, motility (SIM), and growth at 44°C (11). After identification, isolates were cultured in tryptic soy broth (TSB) containing 15% glycerol and were stored at -70°C.
3.2. Evaluation of bap Gene by PCR
Bacterial genomic DNA was extracted from all isolates according to the kit protocol (Sinaclon Co, Iran). bap gene was detected by PCR using specific primers (Table 1). Electrophoresis was performed on 1.5% agarose gels and was visualized by an ultraviolet gel documentation system (BioRad, USA). Acinetobacter baumannii ATCC19606 was used as a positive control.
3.3. Evaluation of bap Gene Expression by Real-time PCR
RNA was extracted from A. baumannii isolates to study the expression of the bap gene. RNA extraction was performed according to the manufacturer's instructions (GeneAll Co., South Korea). The concentration of used RNA was considered to be about 1 - 2 μg. For this purpose, light absorption was measured at a wavelength of 260 nm. Moreover, light absorption at 280/260 nm was assessed to ensure the lack of protein contamination, and light absorption at 260/230 nm was measured to ensure the lack of salt contamination. The cDNA was synthesized from the extracted RNA after DNase I treatment. DNA gyrase A was used as an internal control to study the expression of the bap gene. The sequences of the forward and reverse primer pairs of the two bap and DNA gyrase A genes are shown in Table 1. The temperature program and volume of each material used in the reaction are presented in Table 2.
| Target Gene | Conditions | Volume Reactions |
|---|---|---|
| bap | 1 cycle: 95°C (10 min); 40 cycle: 95°C (20 s), 58°C (40 s), 95°C (15 s), 60°C (30 s), 95°C (15 s) | 2 μL of cDNA, 10 pM of forward and reverse primers for both bap and DNA gyrase A genes, 10 μL of master-mix. H2O up to 25 μL |
3.4. Statistical Analysis
The expression of target genes normalized by housekeeping genes was log2 transformed before analysis. Data were analyzed using a one-way analysis of variance on the linear 2−ΔΔCT dataset and the least significant difference method to analyze the differences between outcome groups. Data were analyzed utilizing the SPSS software version 24 (SPSS Inc., Chicago, IL, USA), and differences were deemed significant where P < 0.05.
4. Results
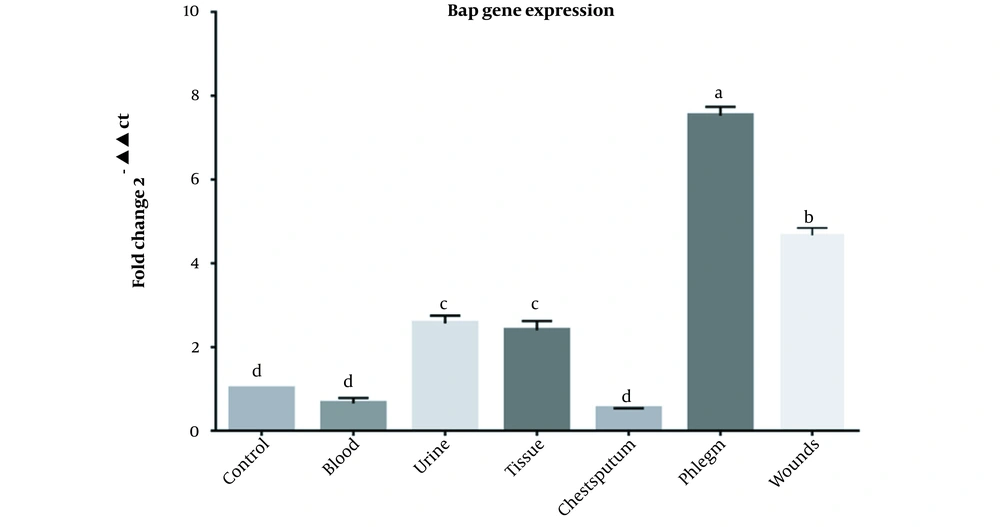
Out of 43 A. baumannii clinical strains from teaching hospitals in Khorramabad, Iran, 23, 8, 3, 3, 3, and 3 specimens were isolated from chest sputum, wounds, urine, tissues and blood respectively. According to the PCR results, all isolates except one had the bap gene. Following the real-time PCR results, 42 isolates expressed the bap gene. The results of real-time PCR for the bap gene of A. baumannii isolates in various clinical samples are presented in Figure 1.
Our findings showed that the relative expression of the bap gene was not significantly different between the control and blood (P = 0.713), sputum and blood (P = 0.997), control and sputum (P = 0.401), while the differences between other groups were significant (P < 0.0001). There was no significant difference in bap expression between urine and tissue groups (P = 0.998), while other groups were significantly different (P < 0.0001). Relative expression of the bap gene in the chest sputum group had the significantly highest value compared to all groups (P < 0.0001). Moreover, the relative expression of the bap gene in the wound group was significantly different from all groups (P < 0.0001) (Figure 1).
5. Discussion
Biofilm formation is one of the most important factors in the pathogenicity of A. baumannii and is effective in bacterial survival in various conditions by binding to substrates (3). For example, biofilm formation in ventilator-associated pneumonia and CAUTIs associated with non-living substrates plays a role in bacterial survival (13). Several factors, including bap protein, are involved in producing biofilms in A. baumannii (14). In the current study, the bap gene was present in all strains except one. Fallah et al. (15) and Mahmoudi Monfared et al. (16) showed that the frequency of the bap gene was 92% and 70.3%, respectively. In the study by Ghasemi et al., the bap gene was detected in 14.2% of A. baumannii isolates, which is not in line with our findings (17). Goh et al. reported a high prevalence of the bap gene (91.7%) in A. baumannii, which is consistent with the results of the present study (18). Ghasemi et al. attributed the difference in the frequency of the bap gene between different studies to the variations in the source and the number of studied isolates. In the research by Ghasemi, 120 A. baumannii was isolated from clinical and environmental samples (17). Therefore, in other studies, the small number of strains and the isolation of strains from clinical samples cause the increasing frequency of the bap gene.
Several studies indicated a strong association between the bap gene and resistance to different classes of antibiotics (15, 19, 20). However, in the current study, the expression of the bap gene was compared between distinct clinical samples, including urine, blood, wounds, tissue, and chest sputum for the first time. Our results confirm that the amount of bap gene expression can also depend on the type of clinical specimen as the highest expression of the bap gene was observed in chest sputum and wound samples and had a significant difference with other samples (P < 0.0001). The latter finding may result from biofilm formation in wound and chest sputum samples more easily than in blood. Chest sputum was collected from a hospitalized patient under a ventilator. Furthermore, our results revealed the need for further investigations on a large number of A. baumannii samples isolated from different clinical and environmental specimens over a more extended period. In addition, the relationships between gene expression and other variables, such as the parts of the hospital and resistance to different classes of antibiotics, need to be evaluated.
5.1. Conclusions
According to the results of the current study, there is a relationship between sample type and the presence of the bap gene, which is one of the main factors in forming biofilms by A. baumannii isolates. Therefore, due to the importance of biofilm in bacterial virulence, detecting the bap gene by molecular assay in hospitalized patients, especially in ventilator-associated pneumonia infections and CAUTIs, which have suitable conditions for biofilm formation, can be helpful in infection control. Considering the prevalence of biofilm-producing A. baumannii isolates and the importance of biofilms in antibiotic resistance, the results could provide a perspective for further research to prevent infections by biofilm-forming A. baumannii strains.