1. Background
The third leading cause of cancer death is gastric cancer (GC) (1). GC-related morbidity and mortality are higher in East Asia, particularly in Korea, Japan, Mongolia, and China, than in most Western countries (2, 3). Tumor metastasis is one of the leading causes of death related to cancer (4, 5).
Tumor metastasis is a complex procedure in which cancer cells proliferate from the original tumor location to other organs. To some extent, metastatic cancer cells retain epithelial properties while displaying mesenchymal properties, including distraction and invasion (6). This biological procedure is related to E-cadherin expression loss and the expression rise of zinc finger E-box binding homeobox 1 (ZEB1), besides up-regulation of Vimentin and N-cadherin (6).
ZEB1 belongs to the ZEB transcription factor family which deals with controlling crucial variables during the invasive front of carcinomas by inducing epithelial-to-mesenchymal transition (EMT), providing cancer cells with a pre-invasive and stem-like phenotype, and determination a much worse clinical prognosis in most human malignancies (7, 8). EMT phenotype and cellular plasticity are induced by ZEB1 (9). ZEB1 increases transcriptional regulation of genes implicated in cancer progression in breast, prostate cancer cells, and GC (10, 11).
Long non-coding RNA (LncRNA) is a kind of non-coding RNA with a length above 200 nucleotides (nt) that can influence target gene expression levels during the transcriptional and posttranscriptional stages (12). Some studies in GC have found that lncRNAs such as GIHCG, (13) ATB, (14) FEZF1 antisense RNA 1 (FEZF1-AS1) (15), SNHG15 (16), and CRNDE (17) are abnormally expressed and linked to cancer progression pathways. The human chromosome 12 gene HOX transcript antisense RNA (HOTAIR) has been shown to serve an oncogenic effect in cancer. HOTAIR dysregulation are linked to cancer progression, including ovarian cancer (18) and colon cancer (19). Nevertheless, there are limited reports on the biological mechanism of HOTAIR in GC (20).
HOTAIR has been identified as an overexpressed oncogene in solid tumors and hematologic cancers. HOTAIR expression was substantially linked with lymph node metastases, TNM stage, and invasion in GC was higher in the tumor than in the neighboring noncancerous tissues (21, 22). In patients with GC, a high HOTAIR expression was further employed to predict poor overall survival (23).
The involvement of HOTAIR in EMT process has been reported in several studies (24). In our previous study, we found that HOTAIR can induce EMT process through its effect on miR200 family members (25). In this study, we hypothesized that HOTAIR has a correlation with the ZEB1, as the target of this family.
2. Objectives
This study aimed to investigate the association between HOTAIR and ZEB1 in GC on the AGS cell line.
3. Methods
3.1. Cell lines and Culture Conditions
The Pasteur Institute of Iran provided the AGS human GC cell line (Tehran, Iran). Cells were full-grown in RPMI 1640 medium (Gibco, USA) complemented with 10% fetal bovine serum (FBS) (Invitrogen, USA) and 1% Pen-Strep (Invitrogen), and then incubated at 37°C, 5% CO2, and saturated humidity. When cells reached a proper confluence, they were passaged with trypsin. An inverted microscope was used to track cell growth. Here, cells were cultivated in the logarithmic growth phase (25).
3.2. Knockdown of HOTAIR in the AGS Cell Line
To silence the HOTAIR gene, AGS cells were transfected by 50 nM of siRNA targeted vs. HOTAIR (siHOTAIR), as well as a negative control siRNA (siNC) obtained from Sigma Company. The siRNAs comprised negative control (MISSION® siRNA Universal Negative Control #1, SIGMA/SIC001) & HOTAIR siRNA–SASI (Hs02_00380445). In 24-well plates, AGS cells were grown to 50% confluence and then transfected by Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The experiment was carried out twice more. At the time of transfection, the plated cells were 70-90 percent confluent (AGS: 60 × 103 cells) and were harvested 48 h later. Finally, real-time polymerase chain reaction (RT-PCR) was used to test the knockdown of the HOTAIR gene, as detailed below (25).
3.3. Assay of Proliferation
An MTT kit (Atocell, England) was previously applied to perform a cell viability experiment based on the manufacturer's instructions. The cell line was seeded on a plate with 96 wells as part of replication research. AGS cells were then transfected with 50 nM of si-HOTAIR, a siNC, and MOCK 24 h after seeding. After 48 h, the Methyl Thiazol Tetrazolium (MTT) solution was added to each well and incubated for 4 h at 37°C in a humidified environment with 5% CO2. Using an Elisa-reader, the wells' absorbance was evaluated at 490 nm (Biorad, USA). Each group had three replicate wells, and experiments were performed three times (25).
3.4. RNA Extraction & cDNA Synthesis from Total RNAs
RNX (CinnaGen) was applied to extract total RNAs from full-grown cells based on the manufacturer's instructions. DNaseI (Sigma) was used to treat the extracted RNA, which was subsequently kept at -80°C. RNA purity was evaluated utilizing a Nanodrop spectrophotometer (Epoch, Biotek-USA) in addition to an A260/280 nm absorbance ratio. The RNA integrity was also tested utilizing electrophoresis on a 2% agarose gel with SafeStain (CinnaGen). Next, using a cDNA Synthesis Kit (TaKaRa, Japan), RNA was reverse transcribed into cDNA following the manufacturer's instructions.
The Oligo dt and random hexamer primers were used for protein-coding genes (ZEB1 and HPRT) and the long non-coding gene HOTAIR. The cDNA was synthesized by a commercial kit (Takara Company), and the final volume of the reaction was made to 10 µL. The reaction was sited for 15 min at 37°C and for 5 sec at 85°C later to stop cDNA synthesis (deactivating the RT enzyme).
3.5. Quantitative Real-time PCR
In the ABI 7100 RT-PCR system (Applied Biosystems, USA), RT-qPCR was done utilizing SYBR-Green PCR Mix (Takara, Japan). The overall volume equaled 20 μL with 1 μL of cDNA, 0.5 μL of the reverse and forward primers (10 μM), and 10 μL of 2x SYBR Green PCR super master mix. For PCR, the conditions were denaturation for 5 min at 95°C, 5-sec 40 cycles at 95°C, and extension/annealing for 30 sec at 60°C. The HPRT housekeeping gene was used as internal control. The Gen Runner & Primer BLAST was used to create primers based on the GenBank cDNA sequences. Table 1 displays the primer sequences. The correctness of the qRT-PCR results was checked by calculating melting curves for the genes plus primer dimer fragments. To finish, genes' expression was evaluated using the Fold change 2-∆Ct technique (Livak technique; [∆Ct: Ct gene of interest-Ct internal control]) in every analyzed sample. Standard curves constructed from raw data were used to measure primers' efficacy (LinRegPcr; Primer efficiency = 2 & Regression = 0.999).
| Target Gene | Primer Sequence (5′→3′) | Forward (F) & Reverse (R) |
|---|---|---|
| HPRT | GGACTTTGCTTTCCTTGGTCAG | F |
| HPRT | GTCAAGGGCATATCCTACAACA | R |
| HOTAIR | GAAAGGTCCTGCTCCGCTTC | F |
| HOTAIR | TCCTCTCGCCGCCGTCTG | R |
| ZEB1 | GCACCTGAAGAGGACCAGAG | F |
| ZEB1 | GGGGTTCTGTATGCAAAGGTG | R |
aHOTAIR, HPRT1, and ZEB1 primers reference: Gen Runner & Primer BLAST
3.6. The Extraction of Total ZEB1 Protein from Cell Line
Protein extraction was performed as previously described (26). In brief, after removing the supernatant and adding some cold PBS, the solution was later centrifuged for 5 min at 1000 g. RIPA lysis buffer containing a suppressor was added to the cell pellet. Next, the suspension was incubated on the ice at 4°C for 30 min. Then specimens were centrifuged for 15 min at 4°C at 13000 g, and the supernatant was collected at -80°C. Protein concentrations were determined using the Bradford method.
3.7. The Evaluation of ZEB1 Protein by ELISA
To this aim, 100 µL of the extracted protein (10 µg/mL) and 100 µL of PBS were added to each of 96 wells and incubated overnight at 4°C. The wells were aspirated and later rinsed twice by a washing buffer, then blocked by adding 300 µL of the inhibitor buffer to each well, and incubated at 37°C for 60 min. Subsequently, after rinsing, 100 µL of ZEB1 antibody (Cusabio, catalog number: CSB-PA026424LA01HU) was added to each well (1 µg per well), and kept for 2 h at RT. The rinsing was repeated. Then, 100 µL of HRP-conjugated secondary anti-rabbit antibody (Sigma Aldrich, catalog number: A6154) was added to the wells at a dilution rate of 1: 10000 and kept at RT for 2 h. Next 100 µL of TMB solution (Sigma-Aldrich, catalog number: T0440) was added to wells and located for 30 min in the dark. After adding 100 µL of H2SO4 stop solution, an ELISA reader was used to read the OD at 450 nm.
3.8. Statistical Analysis
Using one-way analysis of variance (ANOVA) by Graphpad Prism 9 software, statistical analysis was carried out by 95% confidence interval (CI). Data were presented as mean ± SD. A P-value < 0.05 was considered as statistically significant. Analysis of the HOTAIR and ZEB1 expression was done three times for each specimen.
4. Results
4.1. The HOTAIR Expression Level in AGS Downregulated by si-HOTAIR
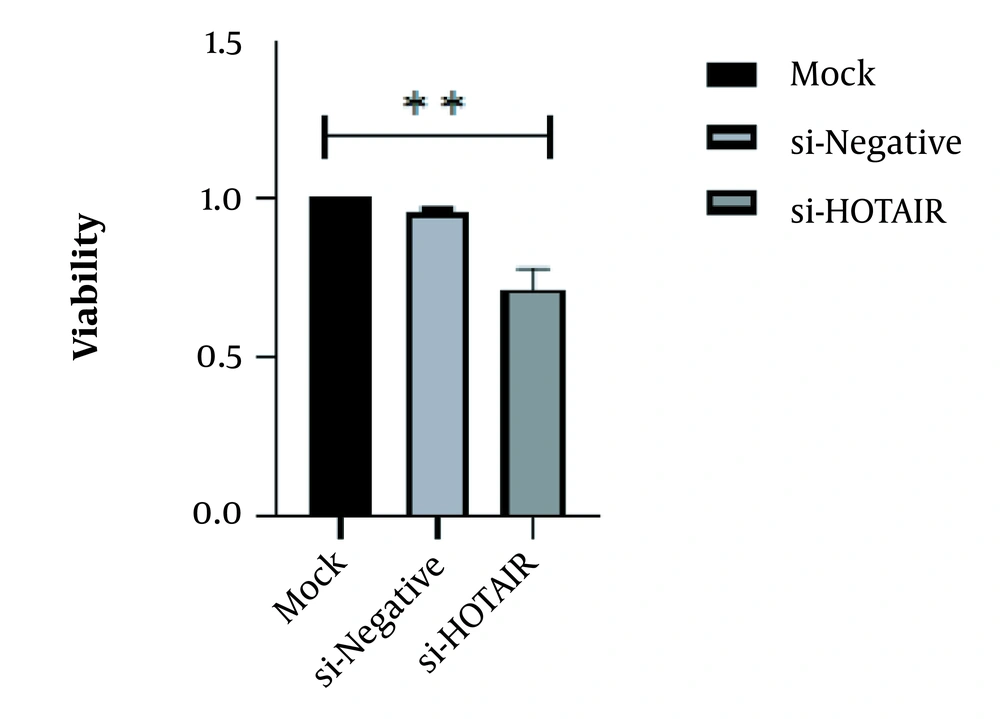
As anticipated, the HOTAIR knockdown resulted in a reduced number of cells in accordance with our previous report on the HOTAIR functional role in GC cell lines (25). A statistically significant decline was observed in developing si-HOTAIR transfected cells as compared with MOCK and siNC groups (approximately for the AGS cell line, Fold change = 0.28; P = 0.0038, **P < 0.01; Figure 1).
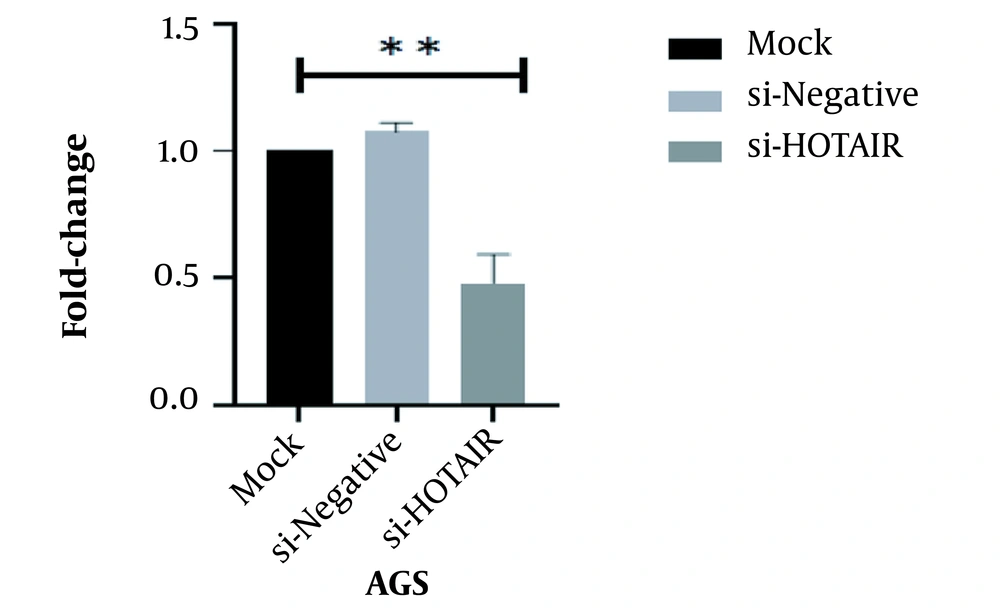
After treating the AGS cell line with siRNA targeting the HOTAIR, the HOTAIR expression level was assessed using quantitative RT-PCR. As illustrated in Figure 2, there was a reduction in the HOTAIR expression level in AGS (Fold change = 0.48; **P < 0.01; Figure 2)
RT-qPCR demonstrated that HOTAIR expression was down-regulated by si-HOTAIR. The HOTAIR expression level in si-HOTAIR-transfected AGS cells. Data were presented as mean ± SD, **P < 0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HOTAIR, Hox transcript antisense RNA.
4.2. The Impact of HOTAIR Knockdown on the ZEB1 Gene Expression
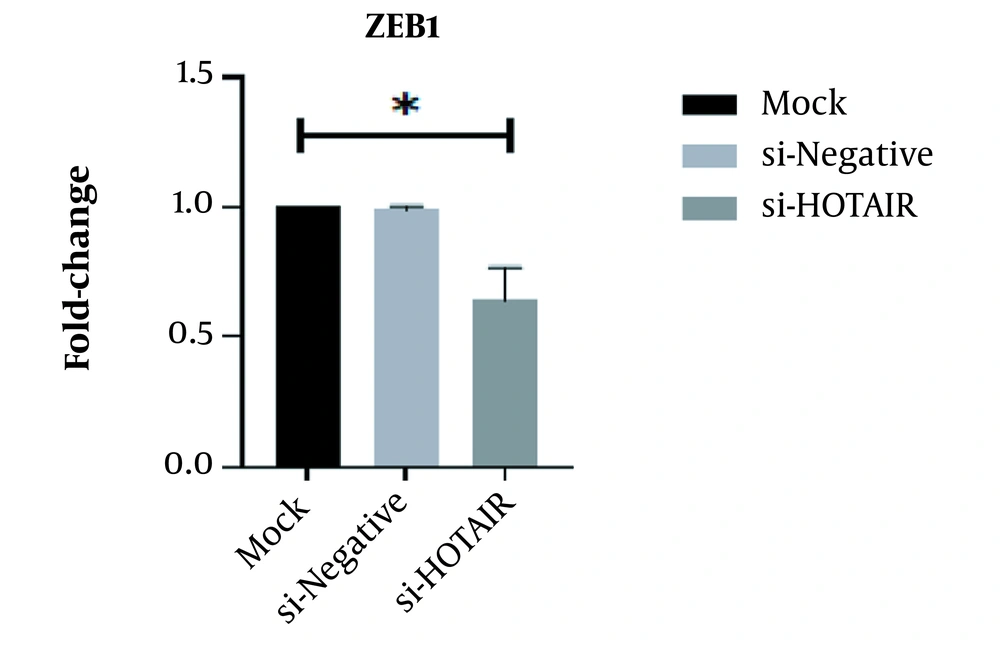
After treating the AGS cell line with siRNA targeting the HOTAIR, the ZEB1 expression level was assessed using quantitative RT-PCR. As observed in Figure 3, the expression of ZEB1 was significantly declined in samples transfected by siHOTAIR. The authenticity of the results was confirmed by one-way ANOVA (Fold change = 0.65; P = 0.0288, *P < 0.05; Figure 3)
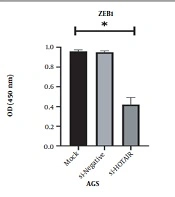
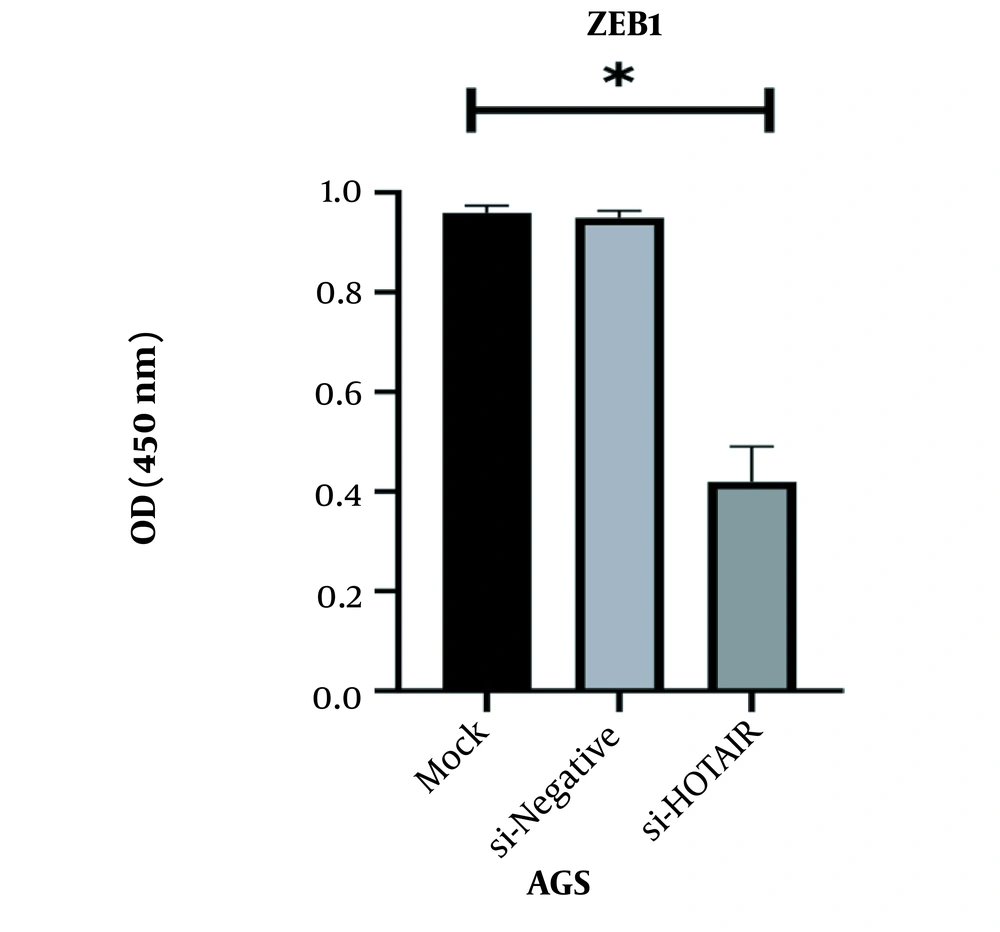
4.3. The Impact of HOTAIR Suppression on the Protein Expression of ZEB1 in AGS Cell Line
The results revealed that protein expression of ZEB1 was significantly reduced in siHOTAIR transfected samples. The suppression of HOTAIR expression was directly associated with the reduction of ZEB1 protein expression. The authenticity of the results was confirmed by one-way ANOVA (P = 0.350, *P < 0.05, Figure 4).
5. Discussion
Studies show that the expression of HOTAIR is significantly higher in tumors compared to normal tissues. On the other hand, many important processes in GC are influenced by this regulatory RNA. Identifying the mode of action of this RNA is effective in better understanding of pivotal molecular mechanisms in GC (27).
The results of this study showed that the HOTAIR gene may have a significant effect on the growth and proliferation of cancer cells. In fact, increased HOTAIR expression can be considered as an important sign in the proliferation and spread of cancerous mass. Regulating the growth and proliferation of cancer cells requires influencing cell cycle and its inhibitors. Considering the mechanism of HOTAIR action on cell cycles and the path of apoptosis can have a significant impact on better understanding the role of HOTAIR on the proliferation and development of malignant tumors. The molecular pathway of HATAIR in GC has also been investigated (28).
Xu et al. stated that the decrease of HOTAIR expression could be accompanied by the decrease of invasion potential and suppressing the EMT pathway. In this study, HOTAIR was shown to impact a wide range of markers and transcription factors involved in the EMT pathway, and the decrease of HOTAIR expression accounted for an important target in suppressing the invasion in GC (23). Zhang et al. also showed that HOTAIR could be regarded as an important factor in the EMT pathway and metastasis of GC (29).
Previous studies demonstrated that lncRNAs are abnormally expressed in cancer and act as oncogenes or tumor suppressors, revealing an important role in cancer prognosis (30). HOTAIR is a possible therapeutic target in treating GC, according to current research findings. HOTAIR has been shown in several investigations to induce EMT by inhibiting target genes (31, 32). We discovered a potential action mechanism for HOTAIR in the up-regulation of ZEB1, one of the most critical markers of EMT.
It was previously revealed that miR-200 acts as a tumor suppressor factor by inhibiting EMT and tumor growth in GC by targeting ZEB1 and ZEB2 (28).
Takei et al. showed that a lncRNA (HOTAIR) is expressed more vastly in 60As6 than HSC-60 cells. Expression of HOTAIR promotes EMT in 60As6 cells. They discovered that HOTAIR binds and targets miR-217, which binds to ZEB1. In the orthotopic tumor mouse model, the knocking down HOTAIR in 60As6 cells greatly declined invasion activity and peritoneal dispersion, and significantly extended longevity. The HOTAIR-miR-217-ZEB1 axis, linked to EMT, appears to prevent peritoneal spread. They concluded that EMT is accelerated in scirrhous gastric tumors by at least two distinct mechanisms: (i) HOTAIR overexpression (60As6 & 44As3 cell lines) and (ii) miR-200 family down-regulation (58As9 cell line). Both methods then work together to increase ZEB1 expression and give cells mesenchymal characteristics (33).
This study revealed that a drop in HOTAIR expression was linked to reducing cancer cell proliferation and growth. The decrease of HOTAIR expression and the ZEB1 gene transcription could happen alongside each other. According to our results, assessing the effect of suppressing HOTAIR expression in the AGS cell line at the level of protein by the ELISA technique showed that suppressing HOTAIR expression could decrease the expression of ZEB1 protein. Considering the key role of the ZEB1 gene as an important marker in progressing the EMT pathway and invasion, it can be concluded that HOTAIR can be a pivotal regulator in the occurrence of metastasis and invasion in GC tumors.
Our results suggested a new mechanism mediated by HOTAIR and ZEB1 that mediates the transitions between EMT and MET programs. As a result of their common molecular properties in the EMT process, ZEB1 and HOTAIR were studied. However, more investigations are needed to define the precise regulation mechanism of HOTAIR on the ZEB1. Further studies are needed to investigate this axis by overexpressing HOTAIR and analyzing interactions between HOTAIR and regulatory elements, evaluation of HOTAIR expression changes on other markers and transcription factors in the EMT pathway, as well as on cell cycle inhibitors and apoptosis for better understanding the effects on cell proliferation and inhibition of apoptosis. Finally, it is recommended to evaluate the effect of HOTAIR knockdown on the expression and translation levels of ZEB1 markers in other cancer cell lines. However, further studies on animal models are necessary.
5.1. Conclusions
The HOTAIR-ZEB1 axis appears to play a key role in human GC, making it a possible therapeutic target for the future. As far as the researchers investigated, this is the first research on the effect of HOTAIR-ZEB1 axis in GC. Nevertheless, more research on the specific mechanism is needed.