1. Background
Chronic myeloid cancer is a type of chronic clonal malignancy that is highly invasive and accounts for about 20% of adult cancers (1). The annual cancer incidence is predicted to be about one to two cases per 100,000 people worldwide (2). A hallmark of chronic myelogenous leukemia is the presence of the Philadelphia chromosome (3). The Philadelphia chromosome is the result of a shift between chromosomes 9 and 22 that eventually fuses the ABL gene on chromosome 9 with the BCR gene on chromosome 22. The resulting BCR-ABL1 fusion is an oncoprotein-encoding gene (4). This activated oncoprotein interferes with proliferation, cell death, and hematopoietic stem cell differentiation (5). Imatinib is the first treatment for these patients. Considered a competitive tyrosine kinase inhibitor, it binds to the oncoprotein BCR-ABL1, suppresses it, and stops signaling (6). However, some patients have shown drug resistance to imatinib due to mutations in the BCR-ABL gene, defects in the expression of drug transporters, or epigenetic changes (7).
Later, information appeared that long non-coding RNAs (LncRNAs) are critical in directing quality expression. Long non-coding RNAs are transcripts of more than 200 nucleotides and are not interpreted into proteins (8). The small nucleus RNA host gene 7 (SNHG7) is a new oncogenic lncRNA that is expressed in various cancers and is associated with the expression of many genes involved in cancer. This oncogenic lncRNA is located at chromosomal position 9q34.3 and is 2176 bp long (9). This lncRNA is the origin of SNORA17 and SNORA43 (10). As of late, numerous considerations have appeared that in numerous tumors and cancers, including colorectal, breast, prostate, and gastric cancers, SNHG7 expression is increased. These studies propose that SNHG7 can be involved in initiation and progression of cancer (11-13). Thus, this lncRNA could be considered a prognostic and diagnostic biomarker and a valuable therapeutic target in cancer.
Fas apoptosis inhibitory molecule 2 (FAIM2), also known as NPM35 and TMBIM2, is located at position q13.12 on chromosome 12. Fas apoptosis inhibitory molecule 2 was discovered when researchers sought to identify anti-apoptotic molecules in the Fas pathway in the resistance created in this pathway in the MRC-5 cell line for lung fibroblasts. After analysis of this protein, it was found that the protein acts as an anti-apoptotic molecule and prevents the induction of cell death by the Fas pathway receptor (14). Studies on colorectal cancer patients also showed a significant increase in the expression of the FAIM2 gene in these patients (15). Research conducted by She et al. showed that the expression of SNHG7 is positively correlated with FAIM2 to elevate tumor proliferation in non‐small cell lung cancer (16). However, the role of SNHG7 and FAIM2 has not been studied in chronic myelogenous leukemia (CML) cancer.
2. Objectives
The current study was designed to assess the expression of SNHG7 in patients with chronic myelogenous leukemia. In this study, for the first time, the expression of this gene in patients with CML who have not received any treatment has been investigated and compared with normal individuals. The results indicate the potential role of SNHG7 and FAIM2 in CML.
3. Methods
3.1. Clinical
30 patients with chronic myeloid leukemia (BCR-ABL +) and 30 normal individuals were selected by confirming their disease status and examining blood cells and karyotype (Figure 1). The selected samples were examined following ethical principles, patient characteristics preservation, and the relevant specialist's approval (Ethical code: EE/1400.3.02.25145/scu.ac.ir).
3.2. RNA Extraction and cDNA Synthesis
After isolation of white blood cells from blood using Lymphodex solution (BAGDIAGNOSTIES), total RNA was extracted using RNX‐ Plus solution (CinaGen, Iran). The quality and quantity of the extracted RNAs were evaluated using gel electrophoresis and a nanodrop device (Thermo SCIENTIFIC). A reverse transcription kit (Amplisense, Russia, cat number: К3-4-100-CE) was then used to synthesize cDNA. This kit contains two RT-mix solutions, RT-G-mix1 and Revertase Enzyme (MMLV). All materials required for reverse transcription, including dNTPs, RNAase inhibitors, random hexamers, and oligo-dT primers, are included in these two solutions. According to the kit instructions, ten µl of RT-mix, 10 µL of RNA, 0.3 µL of RT-G-mix1, and 0.3µL of MMLV were mixed in each tube. The microtubes containing the sample were then placed in a thermocycler for 30 minutes at 37°C according to the temperature-time schedule of the kit.
3.3. RT-PCR and Analysis
RT-PCR reactions were performed using a real-time Rotor-Gene Q (USA) device. Sybergreen (RealQ Plus 2x Master Mix Green Amplicon) was used for PCR. Calculate relative expression levels of RNA was done using Ct values, normalize expression levels of target genes compared to reference genes, and finally compare gene expression data using the Livak (2-ΔCT) method. The primers used in this study were designed using the ensemble database, Primer Blast database, and Oligo software. The sequence of primers is listed in Table 1.
| Gene | Primer |
|---|---|
| SNHG7 | |
| F | TGGTGTGTCCCTTGGTGGAGA |
| R | GGGCTTAGTTACATTGGAGGATTGA |
| FAIM2 | |
| F | GGCGTGCTCTTCGTGCTTC |
| R | TGGCGTCGGTACCCATCA |
| GAPDH | |
| F | ACACCCACTCCTCCACCTTT |
| R | TTACTCCTTGGAGGCCATGT |
The Primers Used in This Study Are SNHG7, FAIM2, and GAPDH Genes
Real-time PCR with the following temperature program was used for both genes: 15 minutes at 95°C and then 40 cycles for 30 seconds at 95°C, 34 seconds at 60°C and 30 seconds at °C. Melting curve analysis, and gel electrophoresis of products was performed to check the specificity of the primers.
3.4. Statistical Analysis
Graphpad version 8 statistical software was used to analyze the data. Mann-Whitney test was used to assess the RNA expression between the patient cases and controls. Data are expressed as SEM. The P-value of less than 5% was considered statistically significant. Additionally, receiver operating characteristics (ROC curves) were used to assess the results quantitatively.
4. Results
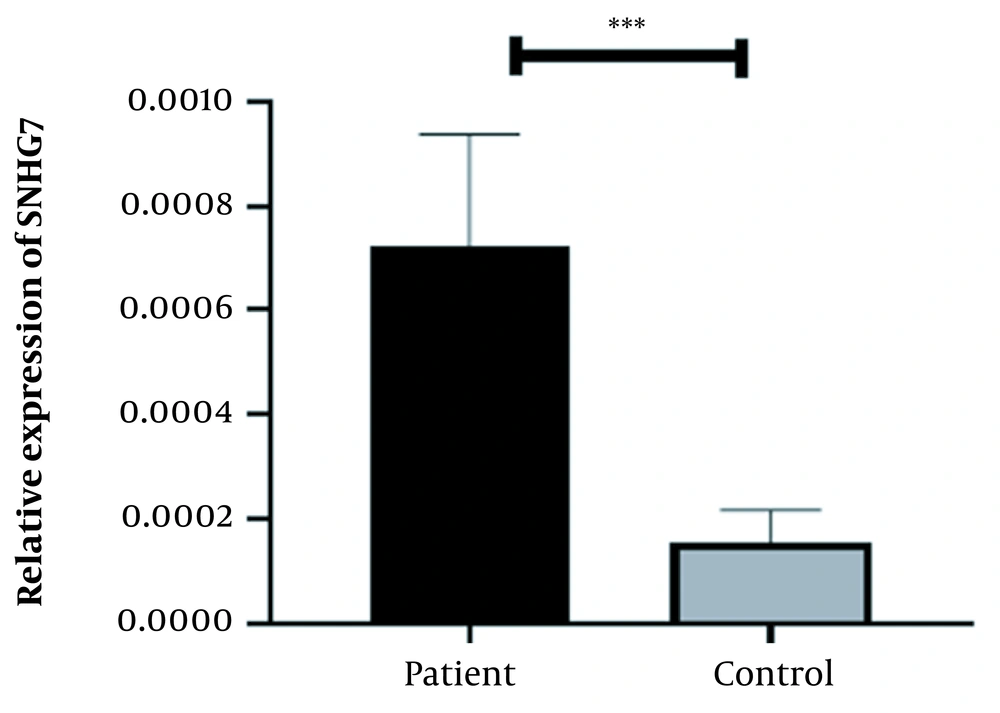
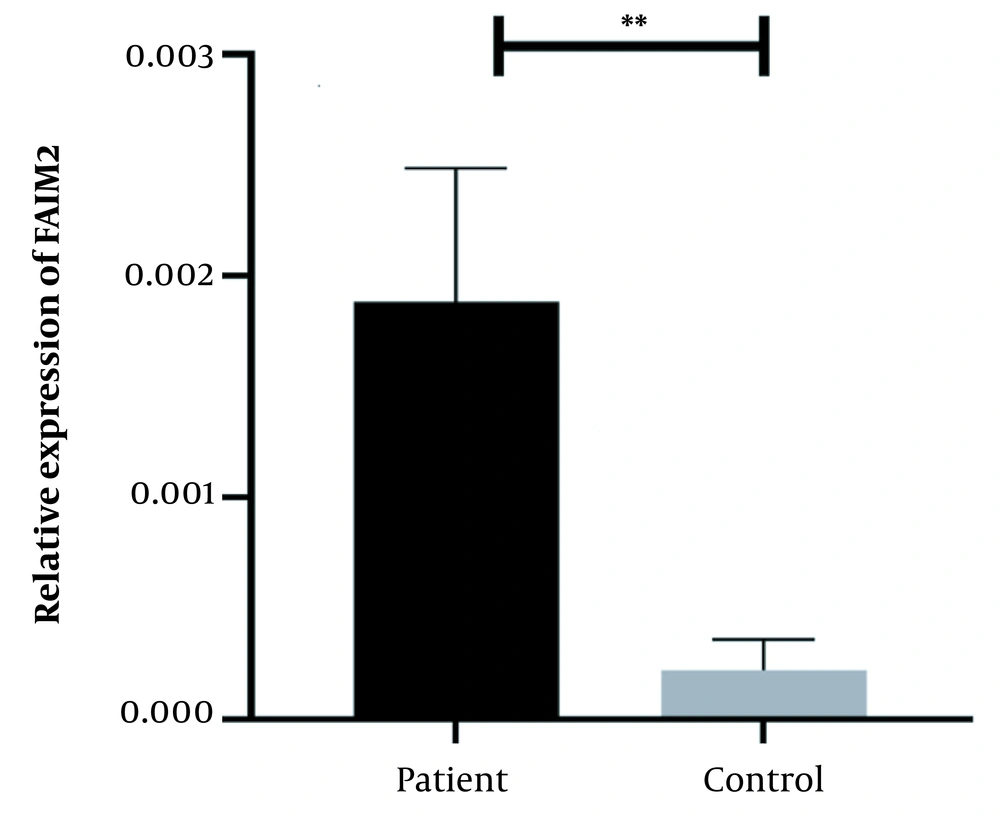
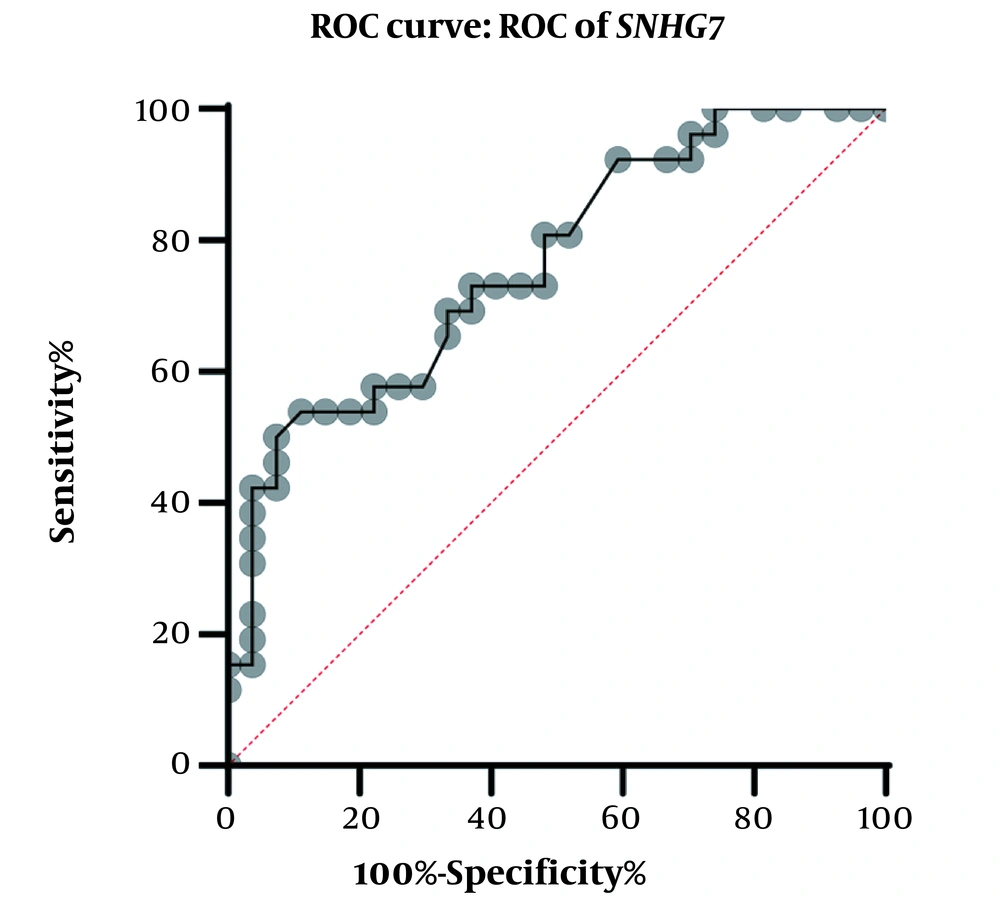
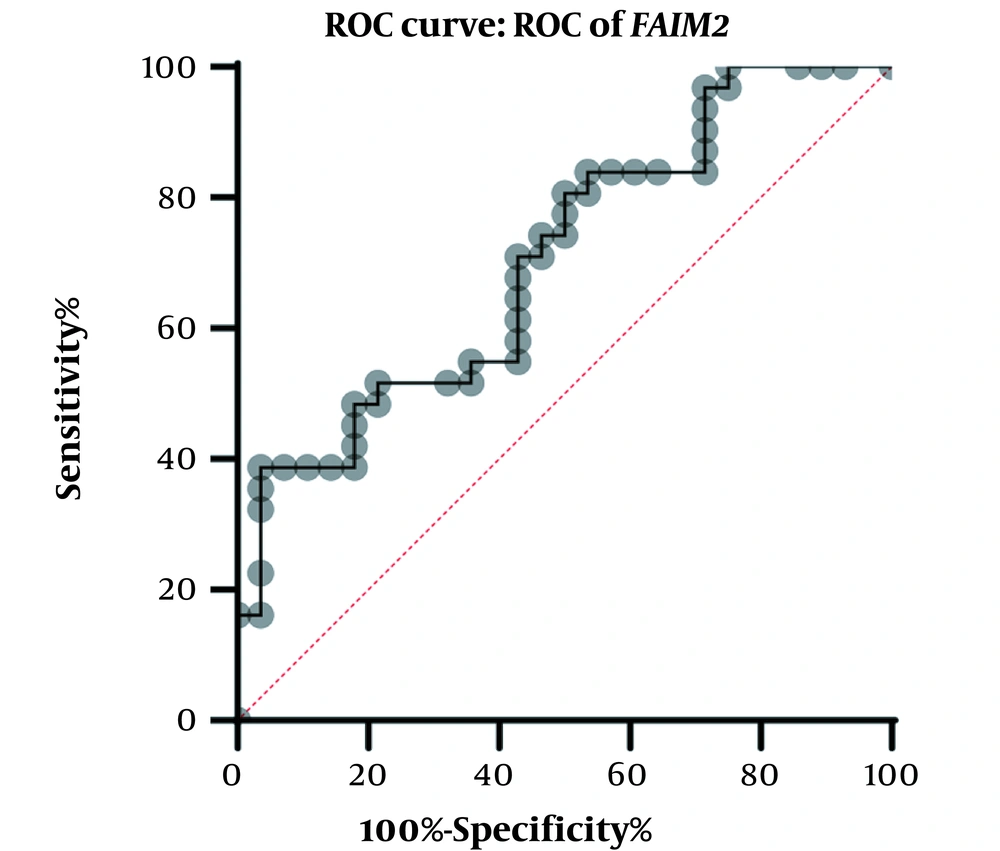
A physician/hematologist evaluated all patients. These samples were taken from people in the age range of 24 to 62. Notably, the patients selected for this study had not received any CML-related drug therapy before registration. Clinical information related to the patient group and the normal groups is given in Table 2. After examining and confirming the patients in terms of karyotype and molecular method of chromosomal fusion detection, the expression level of SNHG7 and FAIM2 in patient samples (n = 30) compared to the control samples (n = 30) was analyzed. In our work, cDNA amplification was performed by real-time PCR of the genes. In addition to the melt curve analysis, the size of the bands was obtained at 168 bp for SNHG7, 173 bp for FAIM2, and 175 bp for GAPDH. The results showed that the expression of the SNHG7 gene (normalized with the expression of internal control gene GAPDH) increased significantly (fold change: 4.5) in patient samples (P < 0.0007) (Figure 2). Furthermore, the expression of the FAIM2 gene (normalized with the expression of internal control gene: GAPDH) between normal and patient groups indicated a significant increase in patients (fold change: 8.5) (P < 0.006) (Figure 3). The results of the ROC curve for the SNHG7 gene (AUC = 0.764, P-value = 0.001) (Figure 4) and FAIM2 gene (AUC = 0.706, P-value = 0.0066) indicate acceptable specificity and sensitivity of the results (Figure 5). The Mann-Whitney test results showed that these genes' expression is much higher in patient samples compared to the normal group. Our data indicate the potential role of these genes in CML cancer. However, the results of this study showed that in chronic myeloid leukemia cancers, we have an increase in the expression of these two genes, which is associated with a positive correlation.
| Group | Age | Gender | Number | WBC |
|---|---|---|---|---|
| Patient | 17 people > 30 years; 13 people < 30 years | 18 men; 12 women | 30 | 21 people > 18000; 10000 < 9 people < 18000 |
| Normal | 19 people > 30 years; 11 people < 30 years | 14 men; 16 women | 30 | All below 10000 |
Specifications of the Patient Samples and Normal Group Used in the Study
5. Discussion
The results of our current study, which evaluated the relationship between the expressions of these two genes in people with chronic myeloid leukemia for the first time, confirmed the previous results. According to the increase in the expression level of SNHG7 and FAIM2 in this study, it is possible that we may encounter similar mechanisms and interactions in chronic myeloid leukemia. However, given that this is the first study in chronic myeloid leukemia, more studies and identification of these genes' upstream and downstream pathways are needed.
Chronic myelogenous leukemia accounts for approximately 12% of all leukemias in the United States and 7 - 20% of all leukemias worldwide (17). The medical improvement of small tyrosine kinase inhibitor molecules (TKIs) during the last decade has shifted the character of CML from a terminal sickness to a persistent sickness. Physicians and their sufferers now have stepped forward with remedy alternatives that permit long-time management of the sickness (18).
The drug therapy using imatinib, which is currently used to treat these patients, is effective and efficient. However, due to the limitations of using in pregnant and lactating women and the drug resistance observed in some people, there is a need to find new methods. BCR-ABL1 fusion, on the other hand, is seen in many patients after TKI cessation, a condition called "untreated recovery" (TFR), which indicates a practical rather than a real treatment (19). Tyrosine kinase inhibitor molecule therapy is far from ideal, and long-term exposure to TKIs with chronic side effects poses a potential health risk and a significant financial burden on the healthcare system. Tyrosine kinase inhibitor molecule intolerance is a recurring clinical problem. Also, some patients develop resistance to TKIs, leading to progressive disease and eventual patient death (20). These concerns highlight the need to discover new strategies for diagnosing and treating CML patients.
Long non-coding RNAs are a class of RNAs with a length of more than 200 nucleotides. These molecules play a regulatory role at the epigenetic, transcriptional, and post-transcriptional levels and play a key role in regulating chromatin structure rearrangement, small RNA processing, apoptosis, and cell cycle regulation (21). Identification of circulating fluid lncRNAs associated with various cancers is well-suited for differential assessments between normal individuals and those with cancer and for identifying early-stage patients with high sensitivity and specificity. In addition, the prognosis of the disease, the risk of tumor metastasis, and the likelihood of recurrence after surgery can be assessed using these biomarkers (22). The importance of the SNHG family in various cancers has been investigated as one of the featured groups of lncRNAs. For example, some studies have shown that SNHG1 is highly expressed in colorectal cancer (23).
In 2020, the team of Shi et al. examined all members of the SNHG family in AML and found that only the expression of SNHG7 and SNHG12 was significantly increased in acute myeloid leukemia (24). On the other hand, studies conducted by the She et al. team in 2016 showed that SNHG7 expression leads to proliferation, metastasis, and invasion of lung cancer cells and, on the other hand, inhibition of cell apoptosis by increasing FAIM2 expression (25).
5.1. Conclusions
The results of previous studies indicate that SNHG7 can enhance cancer cells' proliferation, invasion, and metastasis. It also prevents apoptosis of tumors in the same proportion. Also, an excessive increase in this lncRNA is associated with lower patient survival and more severe pathological complications (11-13). It is possible that this lncRNA could be considered a prognostic and diagnostic biomarker in various cancers and a valuable therapeutic target. As Ziaee et al. study, by qRT PCR, they concluded that the expression level of SNHG7 and FAIM2 was increased in colorectal cancer tissues compared with normal adjacent tissues (15).
Previous studies have shown that SNHG7 interacts with miR193b and is inversely related to its expression. Small nucleus RNA host gene 7 acts as an internal competitive RNA (ceRNA) and binds to miR193b. The expression of FAIM2 is inversely related to miR193b. Finally, it was concluded from this set of studies that SNHG7 binds to miR193b, regulates FAIM2 function, and promotes tumor growth and metastasis. Suppression of FAIM2 gene expression has a similar effect to SNHG7 and stops proliferation, metastasis, and the induction of apoptosis (25). As a result, based on the data obtained from all the studies, it is speculated that SNHG7 induces this effect by regulating the expression of the FAIM2 gene.
Our study shows that the expression of these two genes increased in patients with chronic myeloid leukemia, and more precise identification of these markers will be beneficial to the recovery process of patients or susceptible individuals.