1. Context
In this narrative review, we focused on finding proper genomics and proteomics methods performed in human anthropology and excluded other mathematical techniques.
2. Evidence Acquisition
A hand search with the search query of ((anthropology AND genomics) OR (anthropology AND proteomics)) was implemented in the Elsevier database in title, abstract, and keywords with a total number of 39 results. Subsequently, articles with a publication year equal to or less than 2000 were excluded from the current research. Same strategic search with search query of ((anthropology[Title/Abstract]) AND (genomics[Title/Abstract])) OR ((anthropology[Title/Abstract]) AND (proteomics[Title/Abstract])) was implemented in the PubMed database with total results of 61 articles. The inclusion criteria were: High-impact articles, peer review journals, and relevant subjects in regard to our work. During the screening, 40 duplicates and 5 poorly descriptive articles were found which were not meet our inclusion criteria. Finally, a total number of 55 articles were selected for this narrative review, which contains original and reviews articles as well as one book.
3. Results
Genomics is the study of genes’ properties, including function, evolution, and structure. The first genomics study was performed on bacteria (1). Over time, this discipline has grown and now includes a wide range of research. Genomic studies face several problems when performed on non-living cells containing degraded DNA (2). The idea of implementing genomics in the ancient DNA (aDNA) opens a window to the new molecular techniques in archeological and anthropology sciences (3). Moreover, proteomics, like genomics, consists of a wide range of studies in the field of proteins. Today, with the aid of proteomics, we can study a large variety of proteins (more than 1000) which helps us to acquire valuable information (4).
The science of archaeology deals with the study of past handmade materials, while anthropology deals with the study of human behavior and social interactions. In these two fields of study, researchers use mummies to obtain archaeological and anthropological data (5). In the archeological and anthropological sciences, bones play an important role in characterizing multiple parameters, including age and gender (6). One of the hallmarks to determine the age and gender of the mummies are pelvis and skull volume (7); however, new mathematical methods, the wavelet transform and Fourier transform on the supraorbital margin, and frontal sagittal arcs can be used to determine the gender of human remains using a three-dimensional model of the skull in Geomagic Studio 12.0 software (8). Besides, there are many clues to conducting an anthropology investigation listed as follows.
3.1. Markers in Anthropology and Archeology Sciences
In anthropology and archeology, researchers measure specific markers to obtain the required data for their research rather than performing high-cost methods, including whole genome sequencing or multiple mass spectrometry assessments. Common anthropology and archeology markers are as follows:
(1) Ancestry-informative markers (AIMs) can be used in the genetic aspect of anthropology, and it takes into account multiple population factors, including races and genome admixture. This gene marker has been reported to contain wider sequence variation among different human races (9).
(2) Ancient DNA markers are derived from aDNA, which allows us to study past events like migration. In addition, theories regarding historical events as well as human evolution can be evaluated based on aDNA (10).
(3) Adaptation markers, as the name suggested, signify biological responses to a natural phenomenon with the aim of adaptation to the new environment, which reflects human history (11, 12).
(4) Forensic DNA phenotyping are highlighted for their information provided beyond the human biological state and disclose human phenotypes, clothing, traditions, and other traits with the aid of biological and nonbiological clues (13).
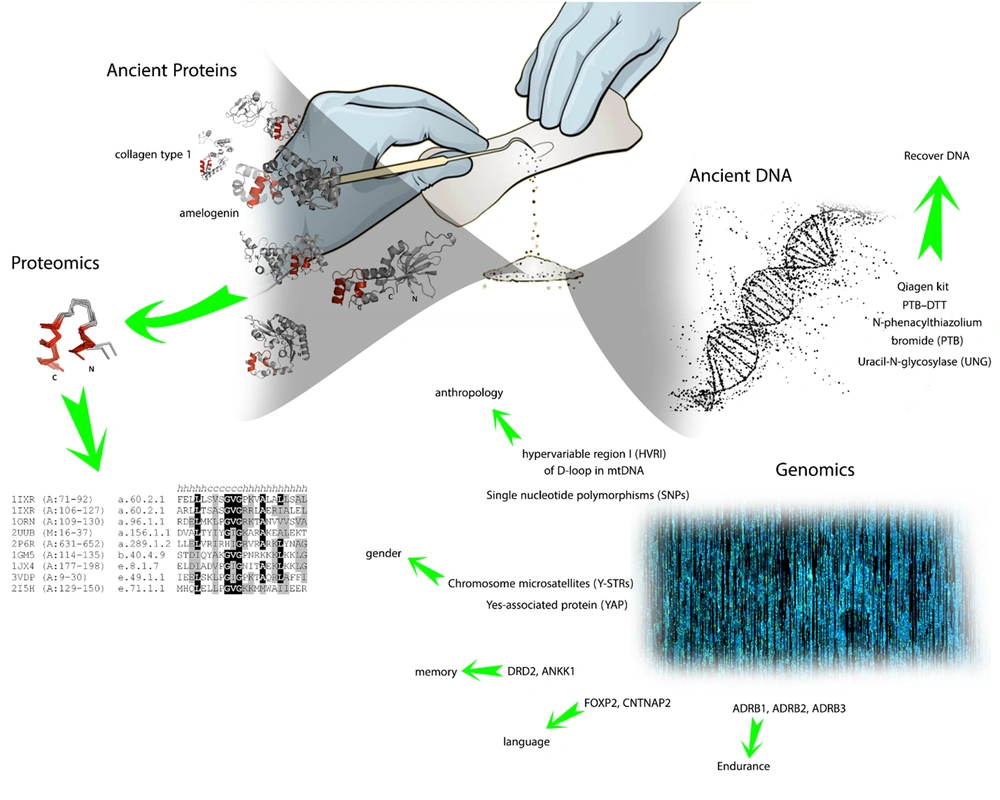
(5) Gene markers provide a wide range of data by dividing genes into particular subunits, which are known to be more expressed in specific areas or actions. Accordingly, we have Hair markers (TCHH, WNT10A, EDAR, SLC24A5, HERC2, TYR, IRF4, SLC4A2, KITLG, LEF1, TYRP1, MC1R, AR/EDA2R, TARDBP, HDAC9, AUTS2, SETBP1, PAX1/ FOXA2, WNT10A, IRF4), circadian cycle (RGS16 (RNASEL), VIP, PER2, HCRTR2 (OX2R), RASD1, PER3 (VAMP3), FBXL3 (CLN5)), ear-related marker (EDN1, FGF3, ABCC11), memory (DRD2, ANKK1), language (FOXP2, CNTNAP2), endurance (ADRB1, ADRB2, ADRB3) (10).
3.2. Post-mortal DNA
In non-living cells, DNA participates in numerous oxidation and hydrolysis reactions, resulting in the cleavage of DNA into smaller fragments (100 - 200 base pairs) (14). In that case, molecular techniques used to detect DNA become useless. DNA fragments encounter a wide range of changes which are characterized by baseless sites, miscoding lesions, and cross-links (interactions between DNA and proteins or sugars) (15, 16). Unlike the former hypothesis, DNA lesions’ site distribution patterns accumulate mainly in the sites named hot spots where repeated sequences are located. These sites can be found in either aDNA or mitochondrial DNA (mtDNA) (17).
Environmental conditions play an important part in DNA degeneration. Hypertonic solutes (like high-concentration sodium chloride) have been stated to decelerate DNA degradation in non-living cells. Nevertheless, even under suitable conditions without hazardous compounds, DNA cannot fully survive for more than 1 million years (15). In the current condition under which humans can live, by aiding kinetic calculations, the studies suggested a maximum length of 100 thousand years for a 100 - 500 DNA fragment to survive, although the mean calculation suggested a much shorter period of 10 thousand years due to hydrolytic damages (18).
3.3. Genomics
We need to cope with DNA changes over time to provide a clear portrayal of past events based on post-mortal DNA. Uracil-N-glycosylase (UNG) is one of the methods used to retrieve post-mortem modification and increase the reliability of results. The mechanism of action of UNG is based on the elimination of deamination products of cytosine from the main sequence. By performing this method, we could also examine the origins of sequence variation (19).
N-phenacylthiazolium bromide (PTB) is another method in which it could overcome intermolecular cross-links caused by enzymes like advanced glycosylation end products (20). The PTB-DTT, and Qiagen kit are the methods to produce high-quality DNA from fragments extracted from specimens; however, PTB-DTT is thought to perform even better quality compared to the Qiagen kit (21). At last, applying a chemical substrate (PTB) to DNA had a desirable outcome in the research on Neanderthal DNA. Chemical treatment continued to be used as a satisfactory alternative to complicated aDNA retrieval methods (22).
Hypervariable region I (HVRI) of the D-loop in mtDNA is a key marker for anthropological and genomics studies. PCR methods are used to amplify short segments into complete sequences (23). Using mtDNA instead of aDNA has several advantages. First, aDNA mostly degenerates into small fragments about 100 bases long, which makes it excruciating work to sequence it completely (24). Second, cytosine (C) is commonly converted to uracil (U) because of deamination, which mimics thymine (T) properties in molecular interaction. Likewise, guanine (G) can convert to adenine (A) in aDNA. These changes are hardly detected by current materials and techniques. Third, aDNA may contain exogenous DNA sources from organisms that live in soil or body microbes (25). This exogenous DNA causes multiple errors in DNA sequencing. Although, damages in aDNA, as mentioned previously, are not randomly distributed and can be identified due to the damage patterns. Current methods in DNA sequencing utilize a DNA library to synthesize short sequences called adapter sequences, which subsequently stick to the end of the short fragments of the sample. Finally, the whole sample can be amplified with primers (26).
Purify aDNA has been long known as a challenging procedure (27). Although novel techniques provided more information about contaminated DNA, the aDNA still contains a tiny portion of the total sample (estimated at 1.3%). With this small value, conducting research on aDNA has multiple challenges (28). Besides, analyzing data accuracy and selecting appropriate DNA sites play important roles in gathering the necessary information for research.
One of the strategies in DNA analysis is to scrutinize sequences for possible mutations, which are known as single nucleotide polymorphisms (SNPs). These sites provide valuable information about population diversity, migration, and admixture. By using these data, we could illustrate a timeline in which the population split occurs (29).
Another site for genomics studies is the human Y chromosome since the haploid state and existence of a wide range of markers make it a potential tool for anthropology studies (30). Population studies are performed at two main sites on the Y chromosome, including Y chromosome microsatellites (Y-STRs) and SNPs. In addition, the Y chromosome contains an abundant transposable element called Alu element and a gene marker named yes-associated protein (YAP), which are useful in population studies (31, 32). Moreover, this chromosome is thought to contain multiple mutations from a long time ago, which makes it a potential site for genomics studies (33, 34).
Microsatellites have long been employed in multiple investigations to prove evidence of illegal actions in the judiciary. Accordingly, various commercial testing kits and multiplexes were developed to facilitate the detection of Y-STRs. Two of them are listed as Powerplex Y 23 by Promega (35) and Y Filer Plus by Thermo Fisher Scientific (36). Before developing Y-STRs detecting kits, anthropology studies were mainly based on conventional methods like SNPs, but nowadays, Y-STRs software packages, including Nevgen and Whit Athey’s Haplogroup Predictor Tool, are widely used for genomics analysis (37).
Besides Y-STRs, other sites may be probed for anthropology and ethnic studies, such as the AZFc region or DYS448, which are evaluated for a possible mutation (38, 39). One of the molecular methods commonly employed in anthropology is PCR multiplex (Genderplex) (40).
After acknowledgment of genetic markers and techniques, it’s noteworthy to consider archaeological methods for DNA extraction from silica materials like bones and teeth, which are commonly used for extracting aDNA (41) (Figure 1). Two main methods are widely known to be applied to mineralization materials, phenol/chloroform/isoamyl-alcohol (PCI) (42) and the spin filter method (SF) (43). Moreover, for DNA extraction in demineralization samples, the QIAamp DNA Blood Maxi kit (Qiagen, Hilden, Germany) is utilized with either PCL or SF methods (44, 45). The final way to acquire aDNA is from Keratinous source materials like hair and skin, although these sources are scarce in post-mortal bodies (46).
3.4. Proteomics
Currently, mass spectrometry is the most efficient method for proteomics studies. However, there are several limitations in performing this method since it demands high-performance instruments and powerful funding resources (47). There are few numbers of proteins that are resistant to the degradation process in non-living cells. In archaeological studies, collagen type 1 (COL1) is commonly extracted and purified from bone for proteomics studies. This protein has a significant amount of hydrogen bonds, and by forming bundles and fibers, it’s resistant to several increasing hazards (48). Another part used in proteomics studies is enamel, which is derived from the tooth. It consists of multiple proteins (up to 10 types). One is called amelogenin, with 2 isoforms encoded at 2 different locations on X and Y chromosomes (49). For proteomics studies, sodium dodecyl sulfate (SDS) is mainly used along with gel electrophoresis. In this method, the protein just deposits a murky sign in electrophoresis, which differs from the well-resolved and distinct bonds of proteins derived from living samples (50, 51).
In post-mortem conditions, proteins cleaved at the carboxyl side of asparagine (Asn) and glutamine (Gln) into multiple fragments (52). Subsequently, these fragments encounter several changes, including deamidation and nonenzymatic reactions (53). Protein samples derived from corpses contain ancient proteins and several contaminants which aren’t thought to participate in deamidation reactions. Since a wide range of proteins is reported to undergo deamination in non-living cells, human proteins could be distinguished from bacteria proteins by measuring the degree of protein deamination (54, 55).
4. Conclusions
DNA strands in a human contain billions of bases. Therefore, it is impossible to analyze all the remaining aDNA bases from the corpus. In this case, researchers perform molecular assessments on a specific number of genes that have been reported to have greater variety among different races. The HVRI sequences in mtDNA and Y-STRs in the Y chromosome are commonly used to distinguish several properties like gender and anthropological variables. Moreover, the AZFc and DYS448 regions of Y-STRs are analyzed to identify the mutation pattern among different tribes. Finally, conducting research on remaining post-mortal proteins provides valuable insights into phenotype variation among different generations. Scientists tend to study proteins that are restricted to the mineral matrix. Since the proteins that exist in bone have higher stability compared to proteins located in the abdomen part, the mineral matrix has a protective role against the degeneration of proteins. This fact highlighted the environmental condition as an inevitable variable for protein protection and genome stability (Figure 1).