1. Background
A pathological change in glandular tissue and abnormalities in milk is caused by bovine mastitis, an inflammation of the udder tissue parenchyma. As a result, the dairy industry around the world suffers significant economic losses (1). In the United States, mastitis-related economic losses are estimated to be USD 2 billion annually (2), with each mastitis case costing producers USD 326 (3). Today, one of the important causes of economic losses in the animal husbandry industry is mastitis caused by coliforms (4). Environmental or coliform mastitis in cattle, the factors involved in which include Escherichia coli, Klebsiella pneumoniae, and Enterobacter aerogenes, often occurs in acute form and is the most common fatal form of mastitis, especially in the case of those dairy cows that are kept in a closed space during the cold season (5). In addition to its effects on dairy cattle, mastitis threatens human health due to the risk of transmitting zoonotic pathogens through ingesting contaminated milk or direct contact with infected cattle (6, 7). The presence of virulent factors such as the cytolethal distending toxin (CDT) gene is related to the pathogenicity of E. coli in the digestive tract and outside it. This gene was isolated for the first time in 1987 from patients with diarrhea caused by E. coli, and its presence has been proven in different pathotypes, such as enteropathogenic E. coli (8). The complete toxin is a heterotrimer consisting of three protein subunits, A, B, and C, coded by three adjacent genes that sometimes overlap each other to a small extent. The expression of all three genes is necessary to produce an active toxin. The cdtB subunit is the active subunit of this toxin, which has deoxyribonuclease activity (9). Subunits A and C are involved in the transfer of subunit B to the target cell, and after entering the target cell, cdtB causes DNA damage. Following the breakdown of DNA connections, irreversible cell cycle arrest occurs in the M/G2 phase, which leads to cell death. So far, five different types of this toxin have been identified in E. coli (CDT I - V). Type 3 is mostly found in animal isolates, and types 1 and 2 are common in human isolates (9, 10).
2. Objectives
The aim of this study was to determine the frequency of CDT genes in E. coli isolates from bovine mastitis in Tabriz city.
3. Methods
3.1. Samples
The present study was performed in the microbiology laboratory at the Islamic Azad university, Maragheh branch, from March to October 2020. In this study, we isolated 100 E. coli strains from 100 cows with acute mastitis in Tabriz city (Northwest of Iran), which had already been identified with biochemical experiments (e.g., Mac Conkey agar, Simmons citrate agar, SIM medium, Triple sugar iron agar (TSI), Methyl red and Voges Proskauer tests (MR-VP), Urea agar, oxidase test, catalase test, and Gram staining).
3.2. DNA Extraction
Escherichia coli DNA was extracted in brain-heart infusion (BHI) agar medium (Merck company, Germany) by boiling at 35°C for 24 hours (11). An Eppendorf tube containing 200 l of sterile TE buffer was filled with 3 - 5 colonies of each sample and thoroughly mixed using a shaker. A boiling ban Marry (100°C) was used to boil the vials for 10 minutes, resulting in boiling water covering two-thirds of the vials. The vials were centrifuged at 9000 g for 5 - 10 minutes for the final step. PCR testing was performed on the supernatant of the DNA vials in sterile Eppendorf tubes. The quality and quantity of DNA extracted were checked using electrophoresis on a 1.5% agarose gel and nanodrop device (Thermo scientific company, Germany).
3.3. PCR Test to Detect Escherichia coli and Intended Genes
In order to confirm the phenotypic diagnosis of E. coli bacteria isolated from bovine mastitis, genotypic diagnosis of the isolates was performed using the genus-specific primer by PCR (polymerase chain reaction) method (Table 1). To detect the tested genes, the PCR method was done in 25 µL, including 12.5 µL of Master mix PCR, 0.5 µL of each specific primer (25 nanomoles) (Table 1), 1 µL (50 ng) of DNA template, and 10.5 µL of double distilled water. The timetable and thermal schedule for each gene are presented in Table 2. The amplified products were run on 1% agarose gel and stained with ethidium bromide (0.5 mg/mL) in a dark room. This study used pathotype STEC O113: NM (Shiga toxin-producing E. coli) as a positive control, and double distilled deionized water was used as the negative control. The PCR product was electrophoresed on 1.5% agarose gel for 1 h.
| Stage | Number of Cycles | Tested Gene Time | Temperature (°C) |
|---|---|---|---|
| 16s rRNA/cdt-I/cdt-III/cdt-IV | |||
| Primary denaturation | 1 | 4′/4′/4′/4′ | 94/94/94/94 |
| Denaturation | 35 | 30′′/30′′/30′′/30′′ | 94 |
| Annealing | 60′′/30′′/30′′/30′′ | 55 | |
| Extension | 60′′/60′′/60′′/60′′ | 72 | |
| Terminal extension | 1 | 5′/5′/5′/5′ | 72 |
4. Results
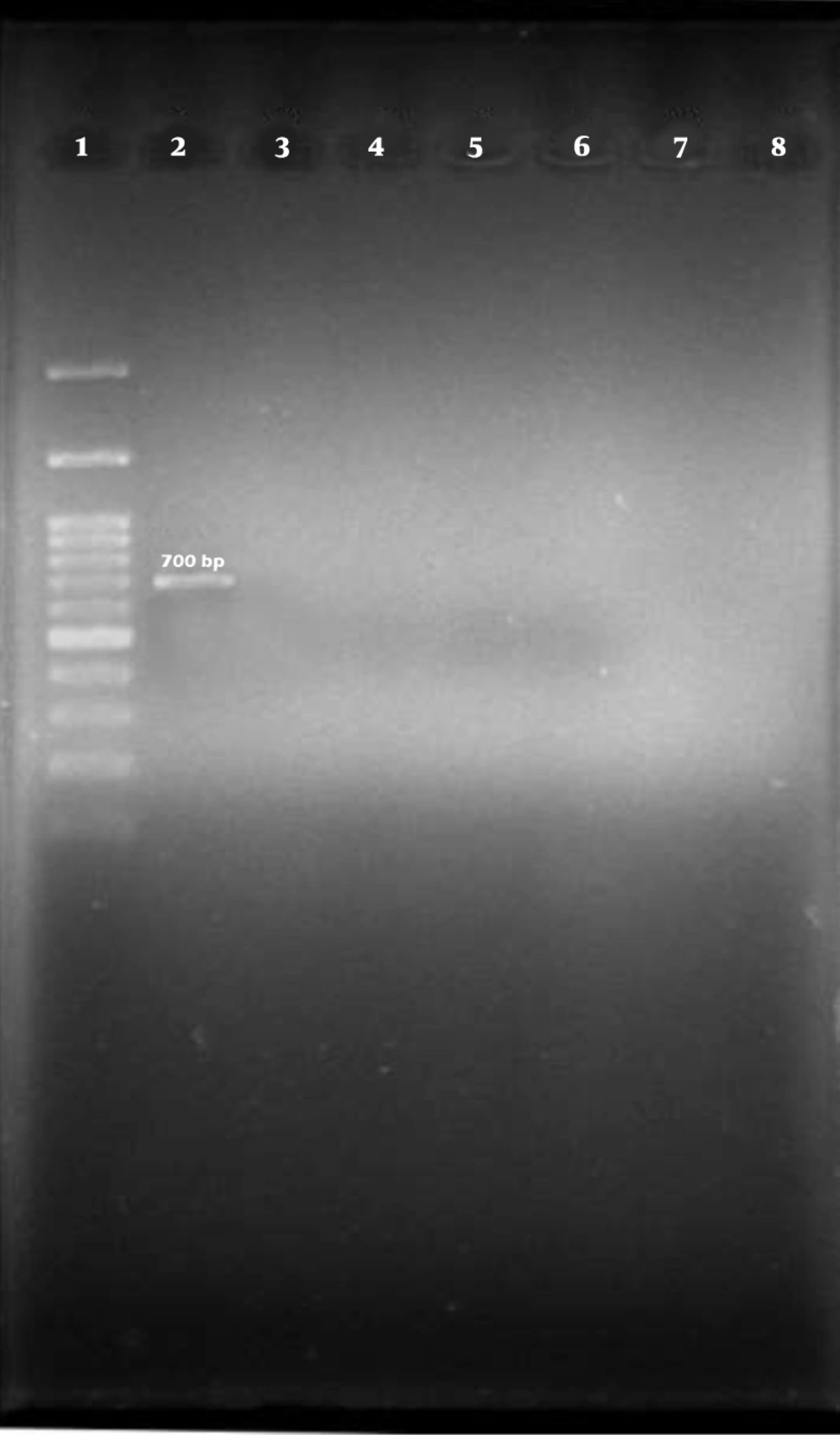
Out of 100 isolates of E. coli that were isolated from bovine mastitis samples by biochemical method and identified by PCR test with specific 16s rRNA primer, 94 isolates were confirmed as E. coli (Figure 1).
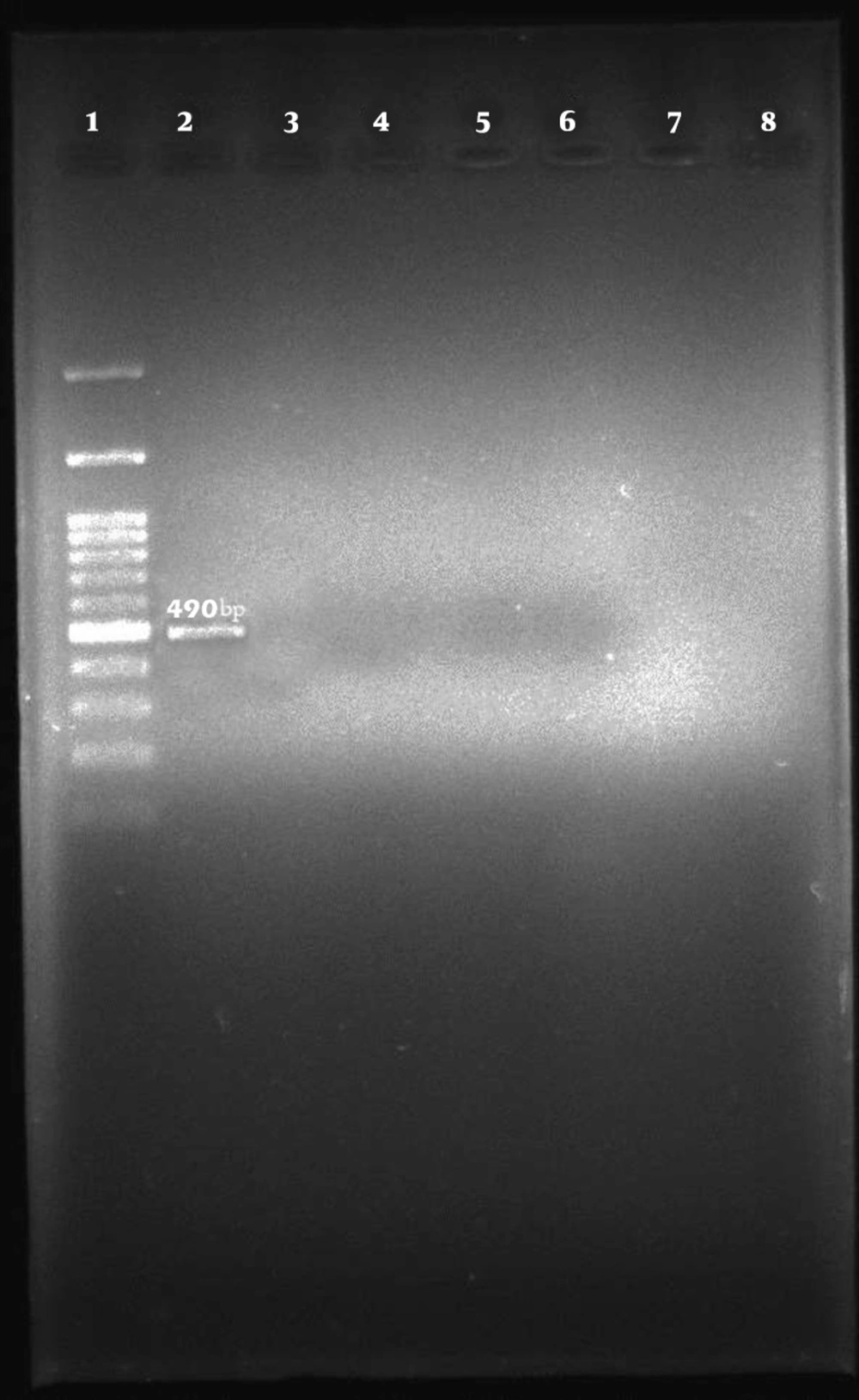
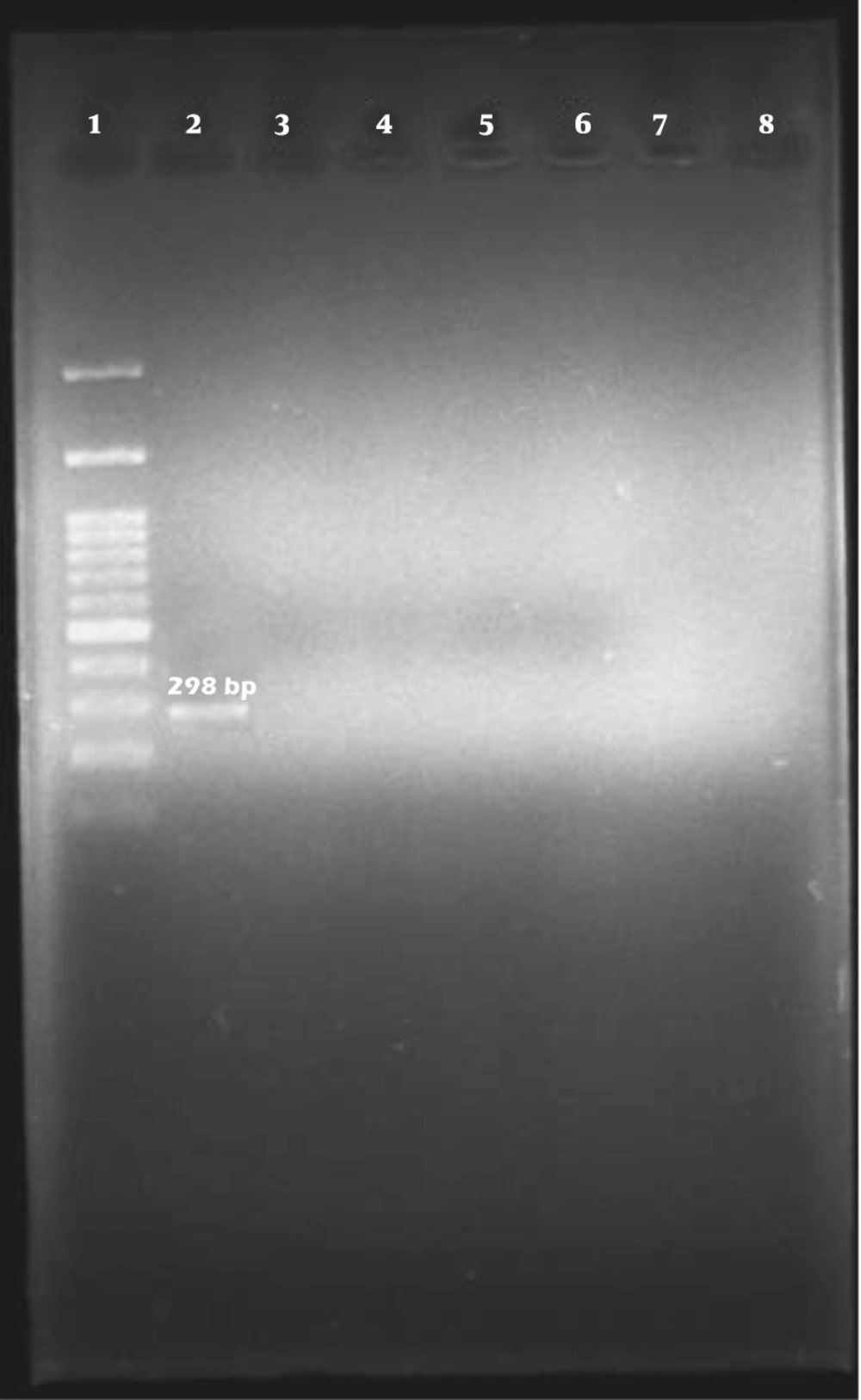
Among the 96 E. coli isolates examined, none of the samples harbored cdt-I, cdt-III, and cdt-IV genes (Figures 2, 3, and 4).
5. Discussion
Environmental health, stress, and livestock immune system are three important factors in coliform mastitis (14), and clinical mastitis occurs most frequently on stressful days after calving (1). Escherichia coli is the most common causative species of coliform mastitis, followed by Klebsiella spp. responsible for most cases of clinical coliform mastitis (15). The importance of CDT toxin-producing E. coli in causing human gastrointestinal and extra-gastrointestinal infections is still unknown. Isolation of CTEC (Cytolethal distending toxin-producing E. coli) strains from patients suffering from septicemia, diarrhea, and urinary tract infections strengthen the hypothesis that these strains are related to human disease (16). Due to the connection of different alleles of this toxin with mobile genetic elements and prophages, its horizontal transfer between different strains is very likely. cdt-III gene is located on a large plasmid containing virulence genes (17), and cdt-I and cdt-IV genes are encoded by lambda prophage (18). Based on the results of this research, out of 100 E. coli isolates identified by biochemical method, 94 samples were identified as E. coli by PCR method and with a specific 16s rRNA primer. This indicates that molecular methods have higher diagnostic sensitivity and accuracy than biochemical phenotypic methods. In the recent study, cdt-I, cdt-III, and cdt-IV genes were not observed in any tested samples. Bouzari et al. reported that the frequency of cdt-I, cdt-III, and cdt-IV genes in patients with diarrhea in Tehran was 45.3%, 52%, and 2.6%, respectively (9). Kafshdouzan and Zahraei Salehi showed that 1.62% of poultry suffering from colibacillosis disease in Tehran had the cdt gene (19). Gomes et al. reported that the frequency of the cdt gene in patients with gastroenteritis, livestock, and food sources in Qatar is 3%, 17%, and 8%, respectively (20). Mainil et al. showed that the frequency of the cdtB-III gene in bovine type 2 necrotoxigenic E. coli (NTEC2) was 83%. Furthermore, they reported that the frequency of cdtB-I and cdtB-IV genes in bovine type 1 necrotoxigenic E. coli (NTEC1) were 3.81% and 4.96%, respectively (21).
5.1. Conclusions
In this study, the frequency of cdt-I, cdt-III, and cdt-IV genes in E. coli isolated from cow mastitis is much lower than in other studies, which indicates the difference in the distribution of the mentioned virulence genes among the different strains. This is probably due to geographical differences and differences in the ecological origin of the isolated strains (food, human, and livestock).