1. Background
The normal function of the thyroid gland is vital for the growth and development of the nervous system as well as growth in childhood and adolescence (1). Thyroid disorders are common, and their diagnosis is based on clinical findings and laboratory and radiological examinations. The major thyroid gland disorders include goiter (diffuse or nodular), hypothyroidism, hyperthyroidism, autoimmune thyroiditis, and thyroid neoplasms (2). Autoimmune thyroid disease (AITD) is the most common cause of acquired thyroid dysfunction, usually manifested as Hashimoto’s thyroiditis or Graves’ disease. Autoimmune thyroid disorders are caused by a combination of genetic and environmental elements and a complex interaction between genetic and environmental factors (3, 4). After puberty, AITD is more prevalent in females than males, while in the premenstrual period, the prevalence is not higher in females (5).
While about 70% of the human genome is estimated to be transcribed into RNA, only 2% of RNAs encode proteins, and the rest are known as non-coding RNAs (ncRNAs). tRNA, microRNAs, long ncRNAs, and rRNA are among ncRNAs (6). Based on the size of ncRNAs, they are divided into two categories: Small ncRNAs (< 200 nt) (> 200 nt) and long ncRNAs (7). Previous studies have demonstrated that there is an association between the increase in long non-coding RNA (lncRNA) expression and the incidence of autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, psoriasis, autoimmune thyroid disease (AITD), and Crohn’s disease (8, 9). LncRNAs can bind to chromatin-altering complexes and direct them to specific genomic sites to induce epigenetic changes and regulate gene expression, including nuclear-enriched abundant transcript 1 (NEAT1) (10).
NEAT1 gene with 4 kb length is located on n 11q13.1. Several studies have reported increased expression of NEAT1 in some autoimmune and inflammatory diseases (11-13). Gene polymorphisms can affect gene expression levels and reduce or increase their function.
2. Objectives
This study investigated the association between NEAT1 rs512715 gene polymorphism and Hashimoto’s thyroiditis and Graves’ disease.
3. Methods
3.1. Subjects and Sample Collection
This case-control study enrolled 133 cases with Hashimoto thyroiditis (HT), 115 with Graves’ disease (GD), and 135 healthy individuals as the control group. Diagnosis confirmation of the autoimmune thyroid disorder was made by a physician based on the results of laboratory profiles and clinical presentations. Patients with other autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, and cancer, were excluded from the study. All patients were informed about the research and consented to participate. Subsequently, 2 mL of peripheral blood was drawn from all participants, collected into anticoagulant EDTA-containing vials, and kept at -20°C for further investigations. The Ethical Committee of the Zahedan University of Medical Sciences approved the study protocol.
3.2. DNA Extraction and Genotyping
The salting-out procedure was employed for genomic DNA extraction from the collected peripheral blood. Subsequently, measurements of the OD 260/280 ratio using a nano-drop instrument were performed to assess the quantity and quality of extracted DNA. Moreover, electrophoresis on 1% agarose gel was done for further confirmation. The DNA samples were stored at -20°C until the genotype analysis. PCR-RFLP method was used for genotyping. The primer sequences, restriction enzymes, and digested fragment lengths are shown in Table 1. The products of PCR were incubated with a specific restriction enzyme, and electrophoresis was performed.
| Gene Polymorphism | PCR Condition | Primer Sequence | Restriction Enzyme | Digested Fragments (bp) |
|---|---|---|---|---|
| NEAT1rs512715 | Denaturation: 950°C (30 S), | F: TCTCTAGGTTTGGCGCTAAACTC | Alu I | CC: 240 + 32 |
| Annealing: 61°C (35 S), | R: GTAACTTTCAGCTGGATGGC | GG:50 + 32 + 188 | ||
| Extension: 72°C (30 S), | CG:240 + 32 + 50 + 188 | |||
| Cycles:35 |
Abbreviations: S, second; F, Forward; R, Reverse.
3.3. Statistical Analysis
Data were analyzed using SPSS-23 software. Categorical and continuous variables were evaluated using the chi-square test (X2) and the independent-sample t-test. A logistic regression method was used to determine the effect of each polymorphism on HT and GD. A P-value less than 0.05 was considered statistically significant.
4. Results
4.1. Demographic Characteristics of Cases and Control Groups
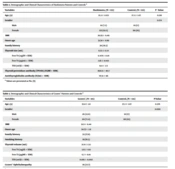
As shown in Tables 2 and 3, this study was performed on 133 cases of Hashimoto’s disease, 115 subjects with Graves’ disease, and 135 healthy individuals as a control group. The mean age of Hashimoto’s patients, Graves, and the control group, was 35.3 ± 1.023, 35.8 ± 1.11, and 37.2 ± 1.07 years, respectively, and there was no significant difference in the study groups regarding age. The frequency of Hashimoto and Graves’ was higher in females than males. The mean age of onset in Hashimoto and Graves’s was 31.34 ± 0.89 and 34.73 ± 1.11 years, respectively. Family history in Hashimoto and Graves’ patients were 29.3% and 27.8%, respectively. Hashimoto and Graves’ patients’ mean body mass index was 26.93 ± 0.49 and 21.9 ± 0.44, respectively.
| Variables | Hashimoto (N = 133) | Control (N = 135) | P-Value |
|---|---|---|---|
| Age (y) | 35.3 ± 1.023 | 37.2 ± 1.07 | 0.136 |
| Gender | 0.174 | ||
| Male | 10 (7.5) | 19 (17) | |
| Female | 123 (92.5) | 116 (83) | |
| BMI | 26.93 ± 0.49 | ||
| Onset age | 31.34 ± 0.89 | ||
| Family history | 39 (29.3) | ||
| Thyroid size (mL) | 8.55 ± 0.54 | ||
| Free T4 (ng/Dl + SEM) | 0.478 ± 0.01 | ||
| Free T3 (pg/mL ± SEM) | 1.16 ± 0.033 | ||
| TSH (mU/L ± SEM) | 62.1 ± 2.5 | ||
| Thyroid peroxidase antibody (TPOAb) (IU/Ml ± SEM) | 460.6 ± 40.7 | ||
| Antithyroglobulin antibody (IU/mL ± SEM) | 717.8 ± 116 |
a Values are presented as No. (%).
| Variables | Graves’ Patients (N = 115) | Control (N = 135) | P-Value |
|---|---|---|---|
| Age (y) | 35.8 ± 1.11 | 37.2 ± 1.07 | 0.376 |
| Gender | 0.099 | ||
| Male | 26 (22.6) | 19 (17) | |
| Female | 89 (77.4) | 116 (83) | |
| BMI | 21.9 ± 0.44 | ||
| Onset age | 34.73 ± 1.11 | ||
| Family history | 32 (27.8) | ||
| Smoking history | 19 (16.5) | ||
| Thyroid volume (mL) | 21.9 ± 1.55 | ||
| Free T4 (ng/dL + SEM) | 3.15 ± 0.11 | ||
| Free T3 (pg/Ml ± SEM) | 6.7 ± 0.19 | ||
| TSH (mU/L ± SEM) | 0.016 ± 0.002 | ||
| Graves’ ophthalmopathy | 26 (22.7) |
a Values are presented as No. (%).
4.2. Frequency of NEAT1 (rs512715) polymorphism in Patients with Hashimoto Thyroiditis and Controls
The frequency of the CC genotype was significantly higher in the HT group than in controls, and the CC genotype may be a risk factor for HT development. The same results were observed in the dominant and recessive models (P < 0.05). Regarding the frequency of alleles, allele C was higher in the HT than in the control group (P < 0.05). More details are demonstrated in Table 4.
| Polymorphism | Inheritance Model | Genotype and Allele | Hashimoto’s Patients (N = 133) | Controls (N = 135) | P-Value | OR (95% CI) |
|---|---|---|---|---|---|---|
| NEAT1 (rs512715) | Codominant | GG | 43 (31.9) | 64 (47.4) | - | 1 |
| CG | 62 (45.9) | 56 (41.5) | 0.064 | 1.6 (0. 97 - 2.8) | ||
| CC | 30 (22.2) | 15 (11.1) | 0.003 | 2.9 (1.4 - 6.1) | ||
| Dominant | GG | 43 (31.9) | 64 (47.4) | - | 1 | |
| CG + CC | 92 (68.1) | 71 (52.6) | 0.009 | 1.9 (1.17 - 3.1) | ||
| Recessive | GG + CG | 105 (77.8) | 120 (88.9) | - | 1 | |
| CC | 30 (22.2) | 15 (11.1) | 0.016 | 2.2 (1.16 - 4.4) | ||
| Over dominant | GG + CC | 73 (54.1) | 79 (58.5) | - | 1 | |
| CG | 62 (45.9) | 56 (41.5) | 0.462 | 1.19 (0.74 - 0.1.9) | ||
| Allele | G | 148 (54.8) | 184 (68.1) | - | 1 | |
| C | 122 (45.2) | 86 (31.9) | 0.001 | 1.7 1.7 (1.2 - 2.5) |
a Values are presented as No. (%).
4.3. Frequency of NEAT1 (rs512715) polymorphism in Graves’ Patients and Controls
The two groups in the Graves’ patients had no significant differences in the NEAT1 rs512715 genotypes and allelic distribution. The results of the genetics models showed similar findings. More details can be seen in Table 5.
| Polymorphism | Inheritance Model | Genotype and Allele | Patients (N = 115) | Controls (N = 135) | P-Value | OR (95% CI) |
|---|---|---|---|---|---|---|
| NEAT1 (rs512715) | Codominant | GG | 49 (42.6) | 64 (47.4) | - | 1 |
| CG | 53 (46) | 56 (41.5) | 0.432 | 1.23 (0.72 - 2.1) | ||
| CC | 13 (11.4) | 15 (11.1) | 0.771 | 1.13 (0.5 - 2.6) | ||
| Dominant | GG | 49 (42.6) | 64 (47.4) | - | 1 | |
| CG + CC | 66 (57.4) | 71 (52.6) | 0.448 | 1.2 (0.73 - 2) | ||
| Recessive | GG + CG | 102 (88.7) | 120 (88.9) | - | 1 | |
| CC | 13 (11.3) | 15 (11.1) | 0.961 | 2.2 (1.16 - 4.4) | ||
| Over dominant | GG + CC | 62 (54) | 79 (58.5) | - | 1 | |
| CG | 53 (46) | 56 (41.5) | 0.464 | 1.2 (0.73 - 2) | ||
| Allele | G | 151 (65.6) | 184 (68.2) | - | 1 | |
| C | 79 (34.4) | 86 (31.8) | 0.617 | 1.11 (0.77 - 1.6) |
a Values are presented as No. (%).
5. Discussion
LncRNA NEAT1 is the primary ligand for estrogen receptors and affects their expression. In malignancies, such as prostate cancer, NEAT1 increases the expression of estrogen receptors (14). LncRNA NEAT1 acts as a transcriptional regulator, and its role as the starting point of tumorigenesis is documented. LncRNA NEAT1 may play a role in gene transcription by cooperating with chromatins and interacting with histones. This lncRNA is associated with increased expression of proto-oncogenes in prostate cancer (15).
There is evidence that NEAT1 is involved in the development of thyroid diseases. NEAT1 probably acts as an oncogene in thyroid cancer; Zeng et al. showed that reducing its expression could suppress thyroid carcinoma (16). A positive effect of NEAT1 signaling was observed on aerobic glycolysis in papillary thyroid cancer cells (17). NEAT1 expression significantly increased in papillary thyroid cancer tissues (18). To our knowledge, the present study is the first report on the role of NEAT1 gene polymorphism in autoimmune thyroid disease. The NEAT1 gene has been identified as a significant regulator of inflammation in some disorders, such as lupus, sepsis, and atherosclerosis. NEAT1 regulates some chemo cytokine levels and inflammasome signaling pathways (19). Li et al. investigated the effect of NEAT1 gene polymorphisms on pulmonary tuberculosis risk. They reported that rs2239895, rs3741384, rs3825071, and rs512715 were unrelated to pulmonary tuberculosis risk (20). The possible role of NEAT1 polymorphisms in other disorders, such as cancer, has been evaluated. Ji et al. concluded that the G > A variant of rs3825071 might confer gastric cancer susceptibility that increases NEAT1 expression (21). Lin et al. investigated the level of NEAT1 gene expression in oral squamous cell cancer (OSCC). The results showed that the expression of NEAT1 in oral cancer cell lines was higher than in normal cells. Increased expression of NEAT1 decreased the survival rate of patients with OSCC. Up-regulation of NEAT1 also reduced the survival rate of OSCC patients treated with chemotherapy and radiotherapy (22). Wang et al. observed that compared with homozygous CC genotype carriers, the GC genotype in the rs2239895 polymorphism was positively associated with the risk of squamous cell lung cancer (23).
In conclusion, the polymorphism rs512715 of the NEAT1 gene could be effective in the development of Hashimoto’s disease, although this effect was not observed in Graves’ disease. The investigated polymorphism seems more prominent in developing Hashimoto’s disease, and Graves’ disease was not affected by these polymorphisms in our study population. Since this is the first study on this polymorphism, more studies on different racial groups are needed to confirm our results.