1. Background
Pseudomonas aeruginosa is considered one of the causes of clinical mastitis with sudden outbreaks in dairy cows. This bacterium affects more than 3% of the herd and causes chronic subclinical mastitis. It causes annually $2 billion economic loss to livestock farmers in America, €93 million in the United Kingdom, and $100 million in Australia (1). Pseudomonas aeruginosa roots cause mastitis in some factors, such as containing contaminated water, improper disinfection of teats, and improper treatment during dry periods. It should be mentioned that this bacterium is probable to be transferred by cow teats (2). Besides its influence on dairy cows, mastitis threatens human health due to the risk of transmission of zoonotic pathogens by drinking contaminated milk or being in contact with infected cows (3, 4). It is the second most common pathogen in surgery, and it is also considered the third most common cause of nosocomial infections after Escherichia coli and Staphylococcus aureus (5). P. aeruginosa is responsible for 11% - 23% of nosocomial infections, especially among patients with cystic fibrosis and people with burns or immunodeficiency (6). One of the important pathogenicity of P. aeruginosa is the secretory system type III, by which some proteins are directly injected into the cytosol of the target cell (7). Five important enzymes, including exoenzymes A, S, T, U, and Y, are transported to the target cell by this system. Exoenzyme T is produced during the release and escape of the pathogen and has ADP-ribosyltransferase activity that is pathologically similar to cholera toxin (8). It is shown that exoenzyme U plays an important role in septic shock intensity and fatality in pneumonia (9). Exoenzyme U is also an active cytotoxin on epithelial cells. It causes lung damage and has a toxic effect on macrophages (10). Exotoxin A is toxic to eukaryotic cells and can inhibit intracellular protein synthesis and damage lung and brain tissues. This toxin also has ADP-ribosyltransferase activity and induces interleukin-1 production, and it has the main role in the pathogenicity of this bacterium (11). The strong potential of Pseudomonas aeruginosa to carry non-chromosomal conjugative genes (on plasmids and transposons) is a good reason for the existence and development of antibiotic resistance, especially when antibiotics are used indiscriminately. Resistance, along with pathogenicity factors, can make this bacteria a potential pathogen.
2. Objectives
This study aimed to investigate the frequency of some important exotoxin genes in P. aeruginosa samples isolated from bovine mastitis in Tabriz City in 2022.
3. Methods
3.1. Samples
This study was conducted from April to November 2022 and included 120 P. aeruginosa strains isolated from 200 cows suffering from acute mastitis in Tabriz, Northwest of Iran. The samples were previously identified through biochemical tests.
3.2. Bacterial Isolation and Identification
The samples were cultured based on MacConkey agar and Eosin methylene blue (EMB) agar and incubated for 24 h at 37°C. The colonies were examined by gram staining biochemical tests like Oxidase, Catalase, Indole, Methyl red (MR), Voges Proskauer (VP), Triple sugar iron agar (TSI), Simmons citrate agar, Arginine dehydrogenase, Ornithine decarboxylase, Oxidation/Fermentation of glucose (OF) and the ability to grow in 42°C (Merck, Germany) (12).
3.3. DNA Extraction
Bacterial DNA was extracted by the boiling method (13). It was conducted on 120 cultured isolates of Pseudomonas aeruginosa on brain heart infusion (BHI) agar (Merck, Germany) at 35°C for 24 h. Then, 3 - 5 colonies from each sample were added to a 1.5 mL Eppendorf tube containing 200 µL of sterile TE buffer and were completely mixed using a shaker. Next, the vials were boiled in a boiling ban marry (100°C) for 10 min till the boiling water level covered two-thirds of the vials. Finally, the vials were centrifuged at 9000 g for 5 - 10 min. The supernatant of vials containing DNA was transferred to sterile Eppendorf for PCR tests. The quantity and quality of extracted DNA were investigated by nano-drop and electrophoresis apparatuses. The extracted DNA was measured by a spectrophotometer at the wavelengths of 260 nm and 280 nm (14).
3.4. PCR Test to Determine Intended Genes
The polymerase chain reaction (PCR) method was conducted in 25 µL, including 11 µL of Master mix PCR, 1 µL of each specific primer (25 nanomoles) (exotoxin A gene primer with 397 bp product: Forward: 5′-GACAACGCCCTCAGCATCACCA-3′ and reverse: 5′-CGCTGGCCCATTCGCTCCAGCG-3′; exotoxin T gene primer with 152 bp product: Forward: 5′-AATCGCCGTCCAACTGCATGCG-3′ and reverse: 5′-TGTTCGCCGAGGTACTGCTC-3′; exotoxin U gene primer with 134 bp Product: forward: 5′-CCGTTGTGGTGCCGTTGAAG-3′ and reverse: 5′-CCAGATGTTCACCGACTCGC-3′) (15, 16), 1 µL (50 ng) of DNA template and 11 µL of double distilled water. Time table and thermal schedule for each gene has been shown in Table 1. The amplified products were run on 1.5% agarose gel and stained with ethidium bromide (0.5 mg/mL) in a dark room.
| Stage | Number of Cycles | Tested Gene Time | Temperature (°C) |
|---|---|---|---|
| A/T/U | |||
| Primary denaturation | 1 | 4′ | 94 |
| Denaturation | 35 | 30” | 94 |
| Annealing | 35 | 45” | 65.8/61/54 |
| Extension | 35 | 60” | 72 |
| Terminal extension | 1 | 5′ | 72 |
4. Results
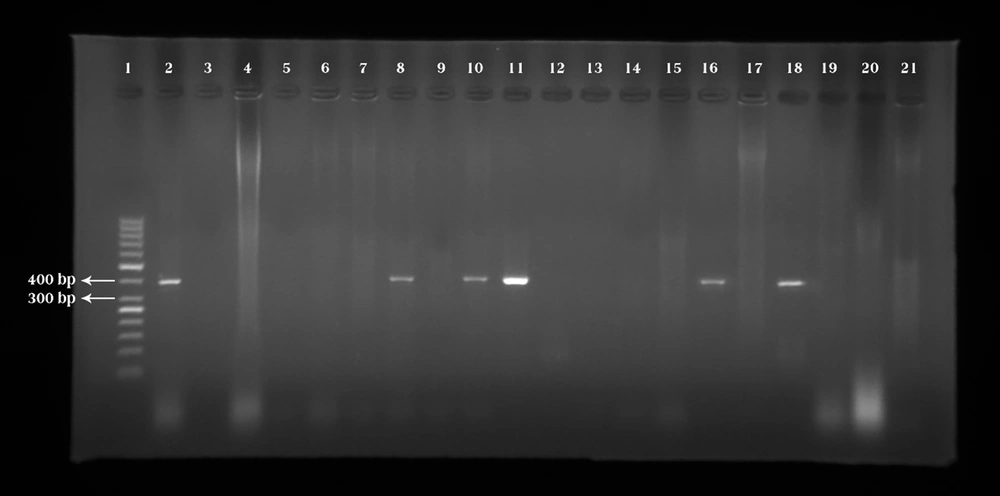
Out of 120 samples isolated from bovine mastitis, 18 (15%) samples of P. aeruginosa harbored the exotoxin A gene (Figure 1).
Lane 1: Marker (50 bp). Lane 2: Pseudomonas aeruginosa ATCC 27853 as the positive control banding within 397 bp, and this is related to the exotoxin A gene. Lane 3: Negative control (Double distilled water). Lanes 8, 10, 11, 16, and 18: Positive samples. Lanes 4-7, 9, 12-15, 17, 19-21: Negative samples
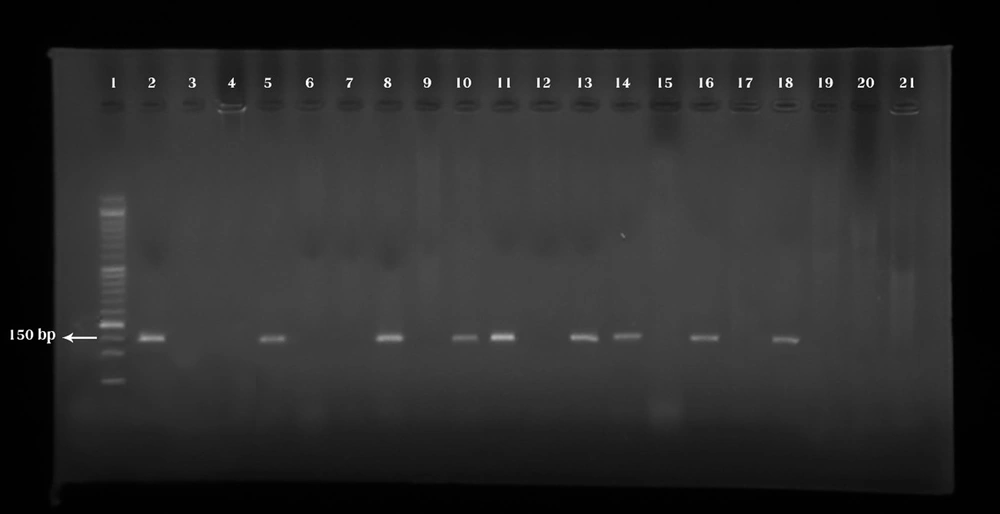
32 (26.6%) samples of P. aeruginosa harbored exotoxin T gene (Figure 2).
Lane 1: Marker (50 bp). Lane 2: Pseudomonas aeruginosa ATCC 27853 as the positive control banding within 152 bp, and this is in connection with the exotoxin T gene. Lane 3: Negative control (Double distilled water). Lanes 5, 8, 10, 11, 13, 14, 16 and 18: Positive samples. Lanes 4, 6, 7, 9, 12, 15, 17, and 19-21: Negative samples.
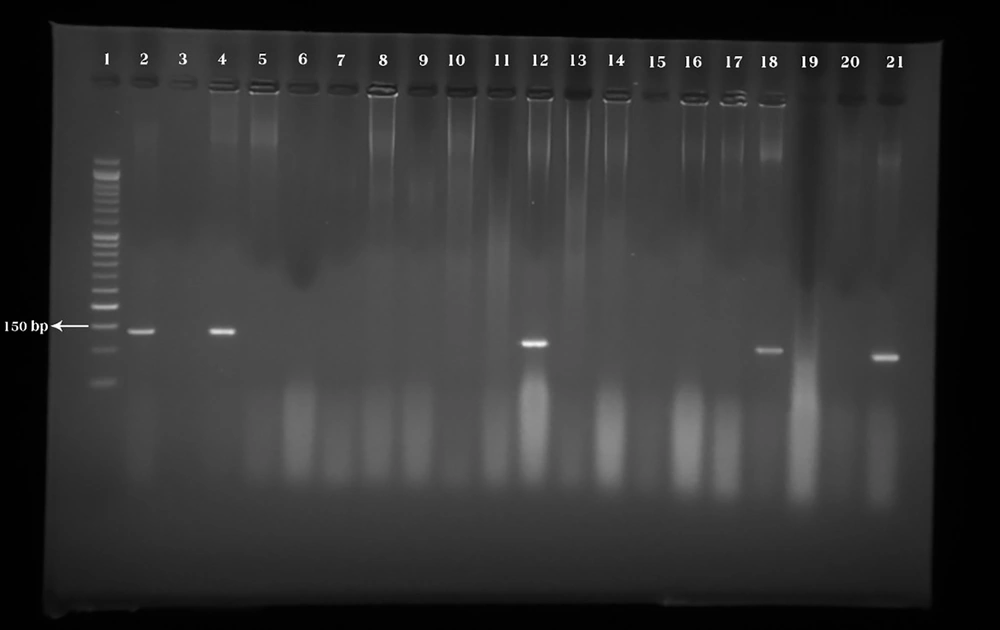
26 (21.6%) samples of P. aeruginosa harbored exotoxin U gene (Figure 3).
Lane 1: Marker (50 bp). Lane 2: Pseudomonas aeruginosa ATCC 27853 as the positive control banding within 134 bp, which is connected with the exotoxin U gene. Lane 3: Negative control (Double distilled water). Lanes 4, 12, 18, and 21: Positive samples. Lanes 5-11, 13-17, 19, and 20: Negative samples
Furthermore, 8 (6.6%) P. aeruginosa samples had all 3 intended genes simultaneously (Table 2), while 76 (63.3%) P. aeruginosa samples lacked the tested genes.
| Exotoxin Genes | Number of Samples | Genes Combination | Number of Samples |
|---|---|---|---|
| A | 4 | A+T | 16 |
| T | 8 | A+U | 8 |
| U | 10 | T+U | 16 |
| None | 76 | A+T+U | 8 |
| Total | 120 |
Based on the chi-square test, the frequency of the genes investigated through exotoxin A, T, and U genes was statistically significant (P< 0.05). In addition, the frequency of the studied genes among gene groups revealed a statistically significant difference (P< 0.05).
5. Discussion
Knowledge about mastitis prevalence, microbial differences, and the risk factors relating to the disease development can significantly improve the prevention and treatment guidelines. Previous studies have shown that mastitis varies among countries as well as farms and cows (17). This meaningful variation in the prevalence of clinical and subclinical mastitis can be associated with ineffective mastitis control programs, environmental factors, and poor hygiene standards in the studied areas (18, 19). The results of the present study revealed that the amounts of exotoxin A, T, and U genes in Pseudomonas aeruginosa samples isolated from bovine mastitis in Tabriz City were 15%, 26.6%, and 21.6%, respectively. This study has been conducted for the first time in Iran on the frequency of important exotoxin genes in P. aeruginosa isolated from bovine mastitis cases. Park et al reported that 82.7% isolates of P. aeruginosa derived from raw milk samples from bovine mastitis had exotoxin U and/or exotoxin S genes containing the invasive (exoU–/ exoS+, 69.4%), cytotoxic (exoU+/exoS–, 8.3%) and cytotoxic/invasive strains (exoU+/ exoS+, 5.0%) (20). Aghaei et al. reported that the rate of exotoxin A, T, and U genes in Pseudomonas aeruginosa samples isolated from clinical samples of hospitalized patients in Qom City (located in the center of Iran) was 100%, 37.9%, and 26.4%, respectively (21). In a study in Poland, it was revealed that the rate of exotoxin A genes in clinical Pseudomonas aeruginosa strains was 76.9% (22). Shi et al. showed that the rate of exotoxin A and U genes in Pseudomonas aeruginosa samples isolated from drinking water and environmental isolates was 92% and 9%, respectively (23). Mahdavi et al. revealed that the prevalence of exotoxin T and U genes among P. aeruginosa isolates from patients in Tehran hospitals were 76.32% and 67.27%, respectively (24). Meaningful differences in the frequency of exotoxin A, T, and U genes in various studies might be associated with different sources of isolation in different areas, their methods of investigation and sensitivity, and also the number and types of samples. The results also show the difference in dispersion of exotoxin A, T, and U genes in Pseudomonas aeruginosa strains; this difference is probably rooted in geographical diversities and also the ecological origin of the isolated strains (foodstuff, human, and different animals).
5.1. Conclusions
The results of the current study showed a relatively high frequency of exotoxin genes in Pseudomonas aeruginosa samples isolated from bovine mastitis. The presence of virulence factors, including exotoxin A, T, and U genes, can be a serious warning to the animal husbandry industry in terms of controlling the disease with this bacterium.