1. Background
Papillomaviruses (HPVs) are a persistent and highly adaptable group of viruses that have evolved in tandem with their hosts to replicate within specific stratified epithelial niches. To date, nearly 450 distinct HPV types have been isolated and sequenced (1). The viral replication cycle of these viruses is intricately linked to the differentiation of the infected epithelium (2). The epidemiology of HPV infections reflects the genetic diversity of HPVs, with specific geographical distributions of different variants of an HPV type (3). This genetic diversity may have implications for the pathophysiology of HPV infection. It is well-established that co-infection with multiple high-risk HPV types is a risk factor for cancer progression, and polymorphisms are associated with an elevated risk of cancer advancement (4). However, understanding the potential methylation profile of human Papillomaviruses (HPVs) as a means to elucidate the purpose of this diversity remains a challenge. Studies have indicated that the methylation of the virus’s DNA is linked to its responsiveness to treatment for the associated pathology (5). An investigation detected increased methylation at 13 CpG sites as the disease progressed, and a high level of methylation was correlated with the risk of CIN2+ in a study examining the relationship between CpG methylation in HPV-16 L1 and persistent infections, as well as the development of cervical carcinoma in Uyghur women (6). These findings suggest that the methylation of CpG sites in HPV could be a valuable predictor of HPV infection persistence and the development of cervical pre-cancer.
2. Objectives
This study aimed to assess the potential utility of the presence or absence of CpG islands in determining the likely oncogenicity of a virus.
3. Methods
3.1. Study Type
This was a retrospective descriptive study.
3.2. Sample Collection Strategy
The sequences for analysis were obtained from pave.niaid.nih.gov (Alphapapillomavirus). A total of 63 HPV genotype sequences were extracted from pave.niaid.nih.gov, a specialized HPV database.
3.3. Selection Criteria for CpG Islands
During the study, any region with a minimum length of 200 base pairs, a GC content of at least 50%, and a CpG ratio of at least 60% were considered as CpG islands.
3.4. Bioinformatics Analysis
Sequence analysis was conducted using dbcat.cgm.ntu.edu.tw, an online methylation analysis tool consisting of 3 components: A CpG Island Finder, a genome query browser, and an analytical tool for methylation microarray data. This analytical tool can process raw data from scanners and identify genes with methylated regions that may impact gene expression regulation.
Statistical Analysis: Statistical analyses were performed using GraphPad Prism v. 8.0.1 for percentages.
4. Results
4.1. Distribution of the Number of CpG Islands
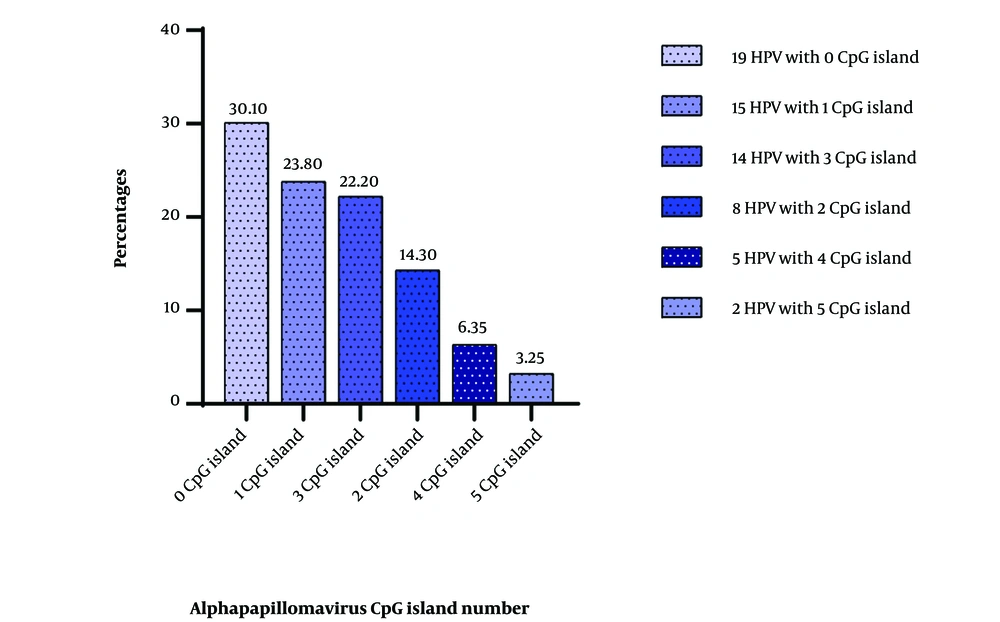
The distribution of the number of CpG islands in all the analyzed genomes is presented in Figure 1 below. It is noteworthy that a significant portion of the encountered viruses lacked CpG islands (30.1%). Conversely, viruses with the highest number of CpG islands constituted only 3.25% of the total number of viruses in the study.
4.2. Profile of HPV Without CpG Islands
The strains listed in Table 1 were found to have no CpG islands when their genomes were examined. Mucosal tropic viruses were found in all of these genotypes. The viruses without detected CpG islands belonged to 5 Alphapapillomavirus species (Alpha HPV 6, Alpha HPV 7, Alpha HPV 9, Alpha HPV 10, Alpha HPV 11). There are no significant types in the Alpha HPV 10 species; these species are generally regarded as low-risk viruses commonly found in condyloma acuminata and laryngeal papillomatosis. There are no major CpG islands in the genomes of subtypes HPV 13, HPV 44, and HPV 73. We found HPV type 16 and 6 subtypes in Alphapapillomavirus 9, which contains the high-risk genotypes responsible for mucosal lesions. These include HPV 31, HPV 33, HPV 35, HPV 52, HPV 58, and HPV 67. HPV 18, which includes 6 subtypes, such as HPV 39, HPV 45, HPV 56, HPV 68, HPV 70, and HPV 97, manifests as Alphapapillomavirus 7, which also has high-risk genotypes often responsible for mucosal lesions. Alphapapillomavirus 11 species and its major type, HPV 34, were also found, as well as Alphapapillomavirus 6 species and its major type, HPV 53. Results indicate the viral genotypes of HPV according to their oncogenic potential, which did not show any CpG islands. It can be seen that high-risk viruses represent about 69% of the viruses without CpG islands in their genomes, while low-risk oncogenic viruses are also observed with a frequency of 15.79%. However, we also observed viruses whose oncogenic risk is not well-defined that showed an absence of CpG islands, notably viruses (67, 97, and 34).
| CpG Island Number and Species | Types Species | Other Papillomavirus Type | Total Number of HPV |
|---|---|---|---|
| 0 | 19 | ||
| 10 | - | HPV 13 (X62843); HPV 44 (U31788); HPV 73 (X94165) | |
| 9 | HPV 16 (K02718) | HPV 31 (J04353); HPV 33 (M12732); HPV 35 (X74476); HPV 52 (X74481); HPV 58 (D90400); HPV 67 (D21208) | |
| 7 | HPV 18 (X05015) | HPV 39 (M62849); HPV 45 (X74479); HPV 56 (X74483); HPV 68 (X67161); HPV 70 (U21941); HPV 97 (DQ080080) | |
| 11 | HPV 34 (X74476) | - | |
| 6 | HPV 53 (X74482) | - | |
| Total | |||
| 5 | 4 | 15 |
4.3. Profile of HPV with a Single CpG Island
Table 2 below shows the different types of viruses encountered that had only 1 CpG island. Indeed, 7 different species of Alphapapillomaviruses were found, which included 5 major and 10 minor Papillomavirus types. A total of 15 viruses had a CpG island; among these genotypes, 46.66% were high-risk (26, 30, 51, 59, 66, 69, 82) and low-risk (6, 7, 11, 32, 43, 54, 91). The vast majority of the CpG islands were identified in the E2 genes in almost 80% of the cases. The size of the CpG clusters ranged from 224 to 410 nucleotides.
| CpG Island Number and Species | Types Species | Other Papillomavirustype | Total Number of HPV |
|---|---|---|---|
| 1 | 15 | ||
| 10 | HPV 6 (X00203) | HPV 11 (M14119) | |
| 8 | HPV 7 (X74463) | HPV 43 (AJ620205); HPV 91 (AF419318) | |
| 5 | HPV 26 (X74472) | HPV 51 (M62877); HPV 69 (AB027020); HPV 82 (AB027021) | |
| 1 | HPV 32 (X74475) | - | |
| 6 | - | HPV 30 (X74474); HPV 66 (U31794) | |
| 13 | HPV 54 (U37488) | - | |
| 7 | - | HPV 59 (X77858); HPV (KR816168) | |
| Total | |||
| 7 | 5 | 10 |
z Abbreviations: LR, low risk; HR, high risk; U, unknown.
4.4. CpG Islands’ Positions on HPV
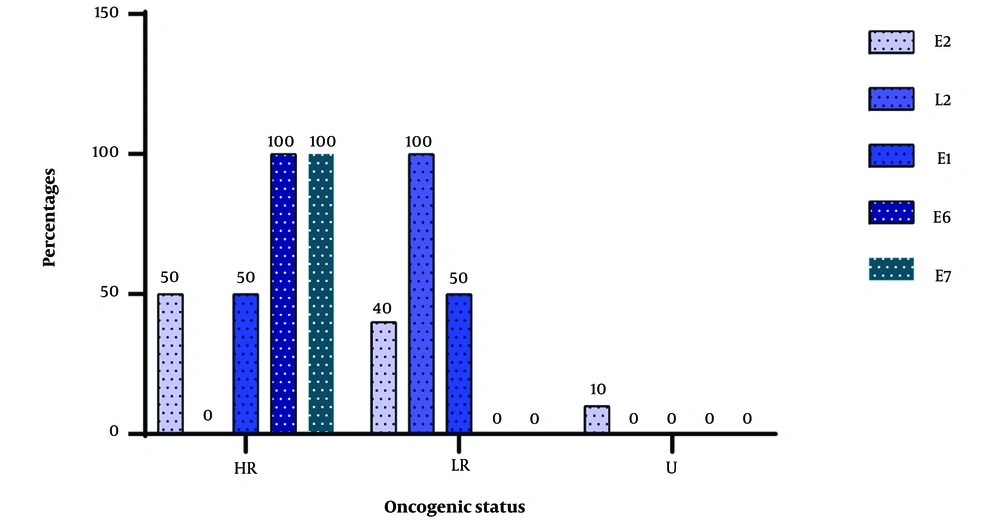
Figure 2 below shows the distribution pattern of the islands for different human Papillomavirus genes. It can be seen that 50% of the CpG islands are located on gene 2 for high-risk genotypes, while for low-risk genotypes, CpG islands were found on gene E2 in 40% of cases. One genotype with unknown oncogenic potential also harbored CpG islands on its E2 gene. No CpG islands were found in the L2 gene of the high-risk genotypes, but the only islands found in the L2 gene for viruses with a single island were found in a low-risk genotype. We were able to identify islands in the E1 gene in equal proportions for both high- and low-risk genotypes. The HPV 30 genotype, which is a high-risk genotype, showed that the CpG island encountered overlapped 3 genes (E6-E7-E1).
4.5. Profile of HPV with 2 CpG Islands
The HPVs presented in Table 3 below exhibit a significant absence of major HPV types. Alphapapillomavirus species 2 had no major types, compared to 3 minor types. HPV species 8, 6, and 3 only showed subtypes of HPV, while HPV species 15 had a major type. Seven types were not clarified (3, 71, 74, 77, 83, 94, 106), and 1 was low-risk (40).
| CpG Island Number and Species | Types Species | Other Papillomavirus Type | Total Number of HPV |
|---|---|---|---|
| 2 | 8 | ||
| 2 | - | HPV 3 (X74462); HPV 77 (Y15175); HPV 94 (AJ620211) | |
| 8 | - | HPV 40 (X74478) | |
| 15 | HPV 71 (AB040456) | - | |
| 6 | - | HPV 74 (U40822) | |
| 3 | - | HPV 83 (AF151983) | |
| 14 | - | HPV 106 (DQ080082) | |
| Total | |||
| 6 | 1 | 7 |
4.6. Position of CpG Islands on Genes for Viruses with 2 Islands
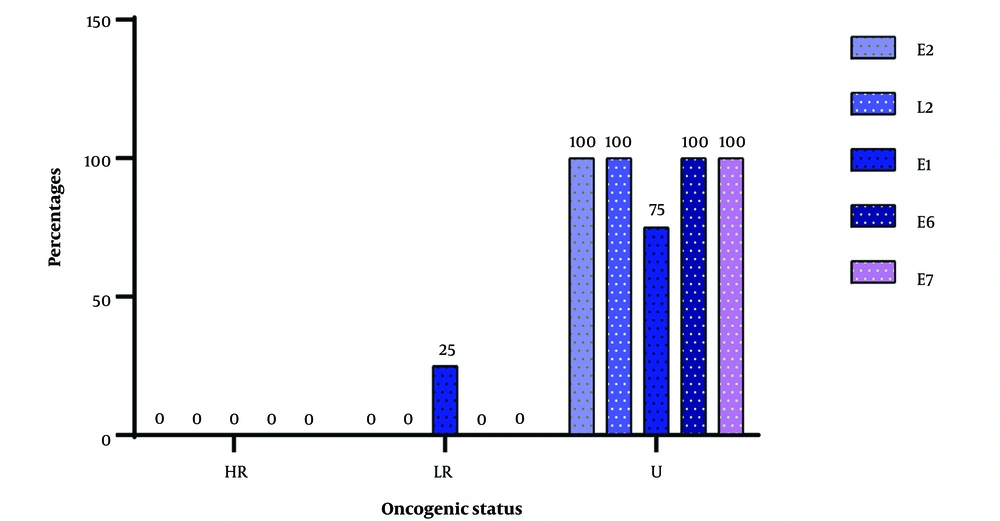
Figure 3 below illustrates the various identification sites of the CpG islands and the viral types in which they were found. There was a complete absence of high-risk viruses among the viruses that had 2 CpG islands, but 1 low-risk virus was present. Almost all the encountered viruses had an unknown oncogenic potential, with nearly 100% of the genes harboring CpG islands, except for the E1 gene, where CpG islands were encountered at a frequency of 75%.
4.7. Profile of HPV with 2 CpG Islands
Table 4 below displays the various Alphapapillomavirus species, in which we identified 3 CpG islands. We can observe that there were 5 species of Alphapapillomavirus that could be identified, with only 2 species presenting major HPV types (HPV 10 and HPV 14). The identified HPV subtypes numbered 12.
| CpG Island Number and Species | Types Species | Other Papillomavirus Type | Total Number of HPV |
|---|---|---|---|
| 3 | 14 | ||
| 2 | HPV 10 (X74465) | HPV 28 (U31783); HPV 29 (U31784); HPV 78 (KC138720); HPV 117 (GQ246950); HPV 125 (FN547152) | |
| 4 | - | HPV 27 (X73373) | |
| 1 | - | HPV 42 (M73236) | |
| 3 | - | HPV 72 (X94164); HPV 81 (AJ620209); HPV 84 (AF293960); HPV 89 (AF436128); HPV 102 (DQ080083) | |
| 14 | HPV 90 (AY057438) | - | |
| Total | |||
| 5 | 2 | 12 |
4.8. Genomic Regions Hosting 3 CpG Islands
A total of 13 viral genotypes were found to contain 3 CpG islands distributed in different viral genes. Among these viruses, we identified 69.23% with unknown oncogenic potential (10, 27, 28, 29, 78, 84, 102, 117, 125) compared to 30.76% of low-risk viruses (42, 72, 81, 89). The range of variation in the CpG islands encountered was from 1470 to 233 nucleotides. No high-risk virus was observed among the viruses with 3 CpG islands.
4.9. Position of CpG Islands on Genes for Viruses with 3 Islands
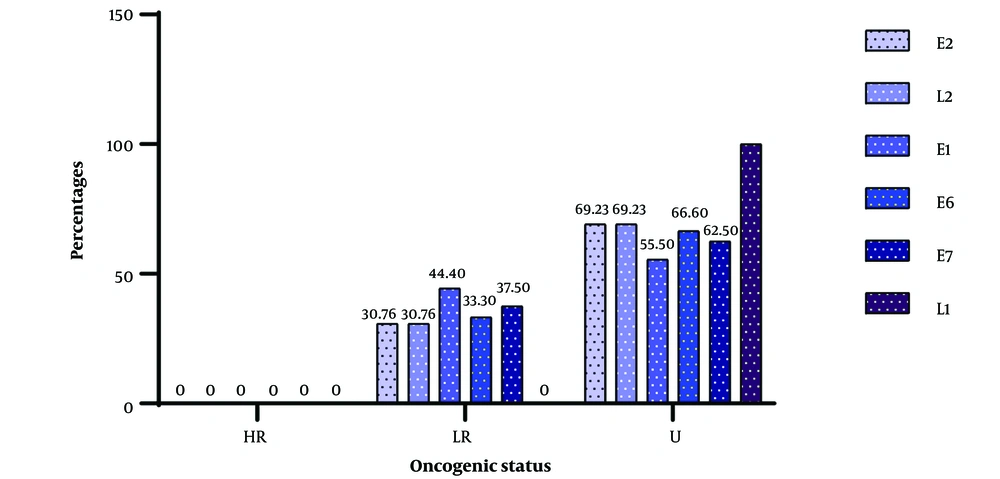
The frequency of the various CpG localization on viruses according to genes is depicted in Figure 4 below. It is clear that genes with low oncogenic risk had fewer than 45% of their genes harboring island CpGs, whereas genes with unknown oncogenic potential had viruses with more than 60% of their genes occupied by island CpGs.
4.10. Profile of HPV with 4 Islands
Table 5 below shows 3 species of HPV, with 1 main type of HPV (HPV2) and 3 secondary HPV types. It can be seen that all of these viruses (100%) were viruses of unknown oncogenic potency. The range of CpG islands observed was between 1247 and 221.
| CpG Island Number and Species | Types Species | Other Papillomavirus Type | Total Number of HPV |
|---|---|---|---|
| 4 | 4 | ||
| 2 | - | HPV 160 (AB745694) | |
| 4 | HPV 2 (X55964) | HPV 57 (X55965) | |
| 3 | - | HPV 114 (GQ244463) | |
| Total | |||
| 3 | 1 | 3 |
4.11. Profile of HPV with 5 CpG Islands
Table 6 below shows the HPV viruses in which 5 CpG islands were observed. Indeed, 2 viruses among those selected showed 5 CpG islands; they are the same species of Alphapapillomavirus, species 3, which presented a main genotype (HPV 61), which has a low oncogenic risk, and a secondary genotype (HPV 62), so the oncogenic power is unknown.
| CpG Island Number and Species | Types Species | Other Papillomavirus type | Total Number of HPV |
|---|---|---|---|
| 5 | 2 | ||
| 3 | HPV 61 (U31793) | HPV 62 (AY395706) | |
| Total | |||
| 1 | 1 | 1 |
5. Discussion
This study aimed to assess the distribution of CpG island profiles on Papillomavirus genomes and comprehend the impact of these predispositions on the neoplastic dynamics of different types of Alphapapillomavirus genus in host cells. Preliminary results indicate that HPV genotypes exhibit epigenomic variations, resulting in different CpG islands in each HPV genotype or subgenotype. This variation may be attributed to different mucocutaneous tropisms of various human Papillomavirus genotypes, potentially influencing the CpG island positions within the genome. These findings align with the work of Chen and colleagues, who observed differences in CpG island profiles in hepatitis B virus (7).
However, some HPVs did not exhibit CpG islands. This absence of CpG islands in certain HPV viruses is predominantly observed in high-risk HPV viruses, suggesting that the absence of CpG islands in these viruses could be a crucial biomarker for their identification. It is also possible that the absence of these CpG islands is a result of the virus’s oncoproteins altering the host environment. Previous research by XIA and subsequent studies by Thomas and Sudhir proposed the hypothesis that genomic methylation may be influenced by host-specific defenses (8, 9).
Regarding their oncogenic potential, high-risk viruses accounted for approximately 69% of viruses lacking CpG islands in their genomes, while low-risk oncogenic viruses accounted for 15.79% of all viruses. This increased absence of CpG islands in high-risk viruses could be a direct consequence of their evolutionary lineage, as demonstrated in a study by Mohita and Perumal, which concluded that CpG depletion among Papillomaviruses and polyomaviruses is linked to the evolutionary lineage of the infected host (10). Silvia and collaborators showed a positive correlation between observed and expected CpG values, with mucosal high-risk (HR) virus types exhibiting the smallest O/E ratios (11).
However, viruses with an undetermined carcinogenic risk, such as HPV 67, HPV 97, and HPV 34, were also found to lack CpG islands. The presence of these viruses with a similar profile to known oncogenic risk viruses suggests their potential oncogenicity within host cells. This observation is logical since genotypes 67 and 97 are subtypes of HPV 16 and HPV 18, which are known to have oncogenic potential. Some authors have isolated these genotypes from high-grade intraepithelial lesions (12, 13).
In some viruses, only 1 CpG island was found, and it was present in 46.66% of cases among high- and low-risk genotypes. In 80% of these cases, the CpG island was predominantly found in the E2 genes. The presence of CpG islands in E2 may result from the regulatory role this gene plays in viral replication. The viral E2 protein, a sequence-specific DNA-binding protein, regulates the Papillomavirus replication cycle and can function as a transcriptional activator or regulator of viral gene expression (14). Several other genes also have CpG islands, indicating the presence of methylation sites within these genes. Given that methylation is a critical phenomenon in the genome, the presence of these CpG islands within these genes serves as tangible evidence of the virus’s regulatory quality, although it is reversible. CpG islands are essential for protecting adjacent housekeeping genes from de novo DNA methylation and maintaining them in a transcriptionally active state (15).
In this study, we found that the size of CpG islands varied with the number of islands encountered. Larger CpG islands were more likely to be found in viruses with multiple islands. This variation may be linked to their oncogenic potential, as suggested by the study conducted by Xue and colleagues, who demonstrated that the lengths and QS heterogeneity of CpG islands differ according to the clinical phases of infection. These data could partly explain the different clinical features observed during various infection phases, warranting further investigation (16).
5.1. Conclusions
This study aimed to assess the distribution of CpG islands in the genomes of Alphapapillomaviruses. It reveals that within this group of Papillomaviruses, CpG islands are notably absent in viruses with a clear oncogenic potential. This pattern of CpG island distribution in viruses could serve as a critical element for identifying oncogenic HPVs. The absence of CpG islands appears to represent an evolutionary characteristic of HPV-related molecular processes. Notably, the more capable an HPV is of causing damage to host tissues (being oncogenic), the fewer CpG islands it possesses. This observation may indeed serve as an evolutionary hallmark for Alphapillomaviruses, which are characterized by the complete absence of CpG islands. The findings of this study could provide a foundational basis for further research on the role of CpG islands in determining HPV oncogenic potential.